BrainVoyager v23.0
VMR to FMR-VTC Space Alignment Using BBR
When working in FMR-VTC space (e.g. for the analysis of sub-millimeter data sets, diffusion data or individual MVPA), coregistration of functional and anatomical data is performed by transforming the anatomical data into functional space as opposed to the normal approach that brings the functional data into native space (and further into ACPC/TAL/MNI space). There are several ways to use BBR when working in FMR-VTC space. The easiest approach is to start with the same first three steps described in topic FA Using Boundary-Based Registration. These three steps include:
- Creation of a whole-cortex mesh in native space from an anatomical data set.
- Running a normal FMR-VMR IA alignment procedure to create the usual IA transformation file as well as creation of a functional volume stored in a near-native space VMR file (new in BV 20.2).
- Running BBR to obtain a fine-tuning adjustment FA spatial transformation file.
Instead of creating, however, a normal VTC file, the produced IA and FA spatial transformation files are used to transform the native-space VMR into FMR-VTC space using the Transform Native VMR To FMR-VTC Space dialog as described in the previous topic VTC Data in FMR Space.
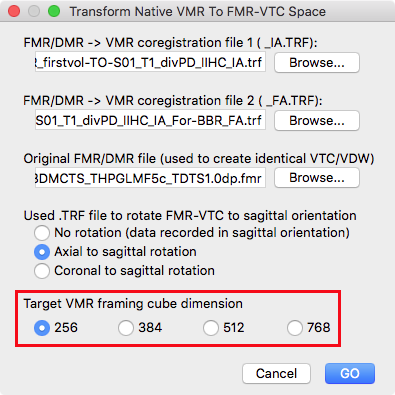
Besides providing the FMR-VMR IA file (step 2 above), the FMR-VMR BBR FA file (step 3 above) and the original FMR file, it is also important to specify the framing cube dimension of the VMR file that hosts the functional data in FMR-VTC space. In the snapshot above, the option 256 is selected in the Target VMR framing cube dimension introduced in BrainVoyager 20.2 (see red rectangle). Selecting the correct target space is important since the dimension of the source (native-space) VMR can be different (in the example data the dimension of the native-space VMR is 384) and only with the correct setting a correct spatial transformation will be calculated and applied when clicking the GO button.
Since BrainVoyager 20.2 the dialog not only applies the spatial transformation to the native-space VMR but it also generates a .TRF file containing s spatial transformation matrix that integrates the IA, FA (if provided) and resolutions of the native VMR and functional data into a single native-space VMR to FMR-VTC spatial transformation matrix. This transformation file can be used to transform any other file from native space to FMR-VTC space, including mesh files. In case that one wants to run BBR in (near) FMR-VTC space, for example, the native-spacer mesh can be transformed using the Mesh Transformations dialog button (see snapshot below).
The generated spatial transformation matrix can be used to transform the native-space cortex mesh (step 1 above) into FMR-VTC space.
Apply obtained spatial transformation (TRF) file to mesh in native VMR space using “Mesh Transformation” dialog. After loading the TRF file ( e.g. “TO_FMR-VTC-Space_3D3D.trf”) apply it normally (forward).
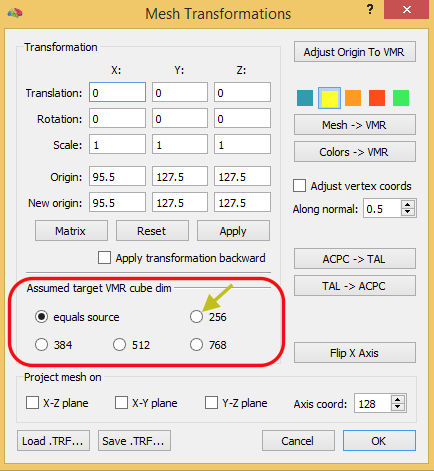
IMPORTANT: As with VMR, specify (new) target cube dim space the same as in 1 (usually 256). This will ensure that scaling works correctly.
Now the mesh should fit to the VMR as well as the FMR-VTC data, which can be checked by projectint the data in the VMR and by creating and saving (2ndary VMR) a functional volume using Show VOL VTC button.
Multi-Run Data. BBR mesh to first run (might be ok if BBR-FA used earlier). Save transformed mesh (e.g. "_Run1.srf". Project mesh in func data of second (third...) run. Run BBR. Use resulting 3D-3D TRF backwards to align second (third) run data to first run using the VTC Spatial Transformation dialog. This will create all VTCs in the same space. More details coming soon...
Copyright © 2023 Rainer Goebel. All rights reserved.