BrainVoyager v23.0
EEG Source Analysis with EEG-fMRI Coupling
Introduction
Exploring the signal coupling of simultaneously recorded EEG and fMRI time-series is attractive because fMRI signals can be automatically predicted (and fMRI patterns can be generated) directly from spatio-temporally selective EEG responses. A cortex-based distributed source EEG model can be used to study "region-specific" EEG-fMRI coupling effects with the EMEG Suite tool. Specifically, it is possible to extract regional power time-courses for fMRI modeling and prediction directly with appropriate options for the Source Analysis (6th tab).
Background
Modeling EEG-fMRI coupling effects entails with the extraction of integrated features from the EEG source responses, e.g. the EEG source power at a given latency or inside a given time or time-frequency interval. The resulting series of "EEG values" is then standardized across all trials and converted to a new time-course at the fMRI temporal resolution (TR). The conversion from this series of EEG measures to an fMRI predictor is performed in two steps: (i) hemodynamic convolution at EEG time resolution and (ii) time-course resampling at fMRI time resolution. The resulting time-course can be used for predicting and modeling, and, hence, localizing the joint fluctuations of EEG and fMRI signals.
Note: Hemodynamic convolution at the original EEG resolution is necessary because the stimulus or any other EEG trigger of interest is normally not synchronized with all fMRI time points, with the sole exception of the fMRI trigger itself. On the other hand, even if the fMRI trigger is used to extract EEG values, the long fMRI repetition time (normally 100:1000 times longer than the EEG sampling period) may suggest using different subintervals within each TR with the result of introducing a time shift between the trigger event and the actual EEG measure.
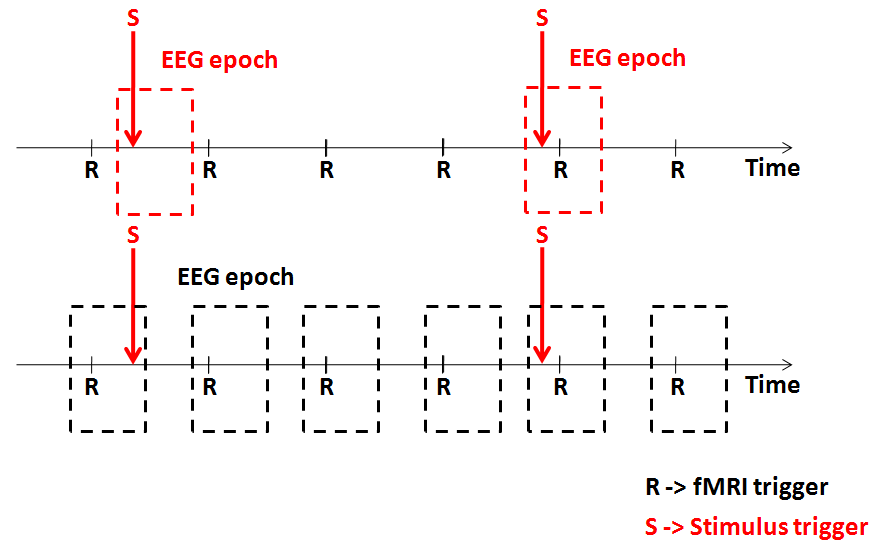
When analyzing simultaneous EEG-fMRI data, there are two alternative approaches for extracting the series of EEG values to be used in the prediction and modeling of the fMRI data: single-trial coupling and continous coupling. Single-trial coupling entails with extracting EEG features from each single-trial response (e.g. the power at a certain latency or inside a given time-frequency box) with respect to the stimulus trigger; these values are assumed to be capable of modulating the amplitude of the event-related fMRI response to that stimulus. Continous coupling entails with integrating the EEG power inside a certain time-frequency box defined with respect to the fMRI trigger. In general, the choice between these two approaches is implicitly driven by the experimental design.
In event-related designs a single EEG trigger of a given protocol condition is associated with the occurrence of single experimental stimulus whose duration is assumed to be zero (i.e. an impulse). Assuming that the absence of a stimulus implies the absence of a neuronal response and, vice versa, the presence of a stimulus implies the presence of the fMRI response, the EEG-fMRI coupling for that condition can be studied directly from single-trial data (STD/TFD) prepared for that specific stimulus.
In block-design experiments a single stimulus trigger only defines the onset or the offset of a block (or, equivalently, the borders of a data segment). In this case, the fMRI trigger should be used and an integrated EEG measure (e.g. the total power in a given frequency band) is calculated for each TR. More in general, this approach is preferred in all cases where EEG-fMRI correlations are studied for long periods corresponding to entire segments rather than short trials of EEG data (e.g. resting-state, free viewing or listening, etc.).
The figure below illustrates the two possible cases of interest.
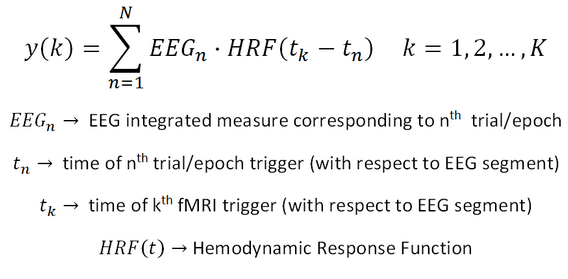
Independently of the approach used (single-trial or continous coupling), the series of EEG values is standardized (i.e. the mean is subtracted and the values are scaled to their standard deviation) and convolved with the double gamma hemodynamic function estimated at the EEG time resolution and resampled at the fMRI time resolution. Assuming a series of N EEG values (corresponding to N EEG epoch-specific measures, either from stimulus or fMRI triggers) and a series of K fMRI values (corresponding to T fMRI time points), the EEG-fMRI coupling time-course can be easily expressed as:
Source Analysis Design Preparation with EEG-fMRI coupling
In order to create EEG-fMRI coupling data from the Source Analyisis (6th tab), before adding the first entry to the table of the source analysis, select the desired input data type (must be single-trial or time-frequency data), make sure to clear the EML list and check the option for EEG-fMRI coupling. Then, add the first entry of the list. For this, besides the inverse solution SMP and the SSM as explained in the Source Analysis chapter, it is now necessary to select the trial timing latency (TRL) file corresponding to the selected data file. Finally, save the new EML list file.
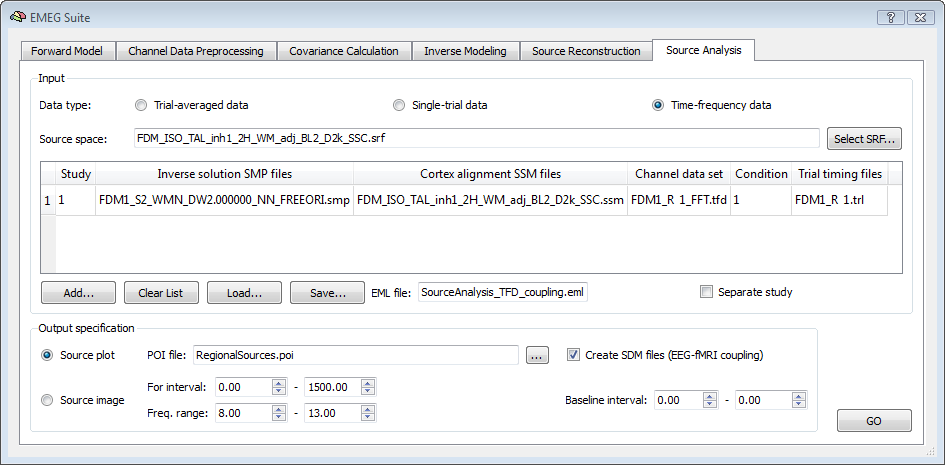
Please note: although the TRL file is saved for each run of the Channel data preprocessing, this is only needed (and asked for) if the EEG-fMRI coupling option is enabled. The TRL file contains the exact timing of occurrence of each trigger of the selected condition with respect to the beginning of the CTC data segment and all trials are included, even if these have been rejected during artifact scanning.
The values in the TRL files are combined with the time of occurrence of the first fMRI scan during the generation of the SDM files (see below). Before starting the analysis, all other required output options must be correctly set. First, a POI file containing the regions to which the EEG data are to projected is to be specified. Second, the time-frequency interval of interest is to be specified. This way, all EEG information in this time-frequency interval with respect to each trigger will integrated and used as EEG measure for EEG-fMRI coupling.
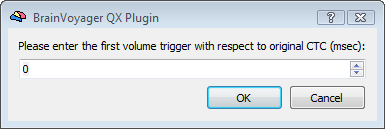
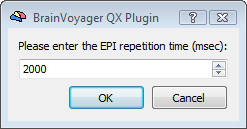
Upon starting the analysis, in order to complete the SDM creation, the program will ask for three important fMRI parameters that are essential for the correct correspondence between EEG and fMRI time points and the application of the hemodynamic correction: (i) the time in millisecond of the first fMRI trigger corresponding to the first fMRI scan included in the analysis, (ii) the fMRI repetition time (TR) and the number of scans constituing the fMRI time-series.

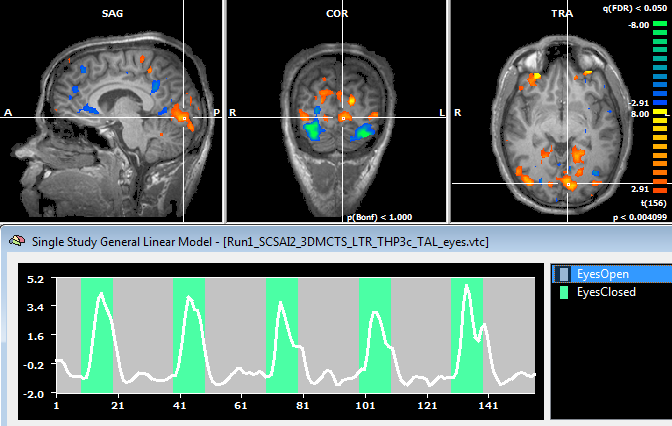
At the end of the run a new SDM file will be created for each region in the POI list file. The name of the SDM file will be include the name of the input data file, the name of the patch and an indication of the time-frequency box to which the EEG integrate measure is referred. This file can be directly used for a GLM analysis of the fMRI data both in the whole-brain volume (VTC) and in the cortex mesh (MTC).
References
Esposito, F., Mulert, C., Goebel, R., 2009. Combined distributed source and single-trial EEG-fMRI modeling: application to effortful decision making processes. Neuroimage. 2009 Aug 1;47(1):112-21.
Esposito, F., Aragri, A., Piccoli, T., Tedeschi, G., Goebel, R., Di Salle, F. Distributed analysis of simultaneous EEG-fMRI time-series: modeling and interpretation issues. Magn Reson Imaging. 2009 Oct; 27(8) :1120-30.
De Martino F, Valente G, de Borst AW, Esposito F, Roebroeck A, Goebel R, Formisano E. Multimodal imaging: an evaluation of univariate and multivariate methods for simultaneous EEG/fMRI. Magn Reson Imaging. 2010 Oct;28(8):1104-12.
Copyright © 2023 Fabrizio Esposito and Rainer Goebel. All rights reserved.