BrainVoyager v23.0
Functionally Informed Cortex-Based Alignment
Introduction
Curvature-driven cortex-based alignment (CBA) achieves good corresondance of macro-anatomical landmarks (sulci, gyri) across subjects that also leads often to better alignement of functionally defined areas. While improved alignment of functional areas will usually lead to improved vertex-wise statistical results of multi-subject whole-cortex fMRI analyses, the remaining residual variability of functional areas after CBA will still lead to less statistical power as compared to an explicit ROI-alignment approach. In order to address this issue, functionally informed CBA (fCBA) uses functional information of corresponding regions from a separate localizer scan to drive alignment in addition to the minimization of macroanatomical variability across subjects. By using proper weighting of anatomical and functional forces, fCBA creates a group aligned space with strong spatial correspondence of functional regions while maintaining a high level of anatomical fidelity. In case that corresponding ROIs are available from functional localizer runs for all included subjects, fCBA allows to perform a single main analysis that integrates whole-cortex and ROI-based analysis as compared to the usual approach that involves running two separate analyses, i.e. one analysis for whole-cortex analysis and a separate analysis for aligned ROIs.
It is important to note that fCBA should not be used when the goal of a study is the assessment of functional variability of corresponding regions across subjects (Frost & Goebel, 2012). FCBA is recommended in cases where one wants to optimize group-level statistical analyses in the same way as one usually would use ROI-aligned multi-subject analyses (Frost & Goebel, 2013).
CBA Informed by Anatomical Landmarks
Note that fCBA can also be used to include macroanatomical landmarks as explicit targets for alignment; in that case corresponding patches-of-interest (or surface maps) are not resulting from functional localizer runs but are derived from anatomical information, e.g. by labelling corresponding anatomical landmarks in individual cortical hemispheres. An example of such an anatomical application of fCBA is the explicit inclusion of the mid-fusiform sulcus (MFS) for a vsiual area atlas (Rosenke et al, in preparation) in order to better align functional areas in the vicinity of MFS.
Background
It has been demonstrated that, after curvature driven cortex based alignment (CBA), macroantomical variability between subjects is substantially reduced and indeed almost completely removed for major sulci. Furthermore as a consequence of increased macroanatomical correspondence, a concomitant increase in the overlap of a set of investigated functional areas across subjects was observed (Frost and Goebel, 2012). The gain in overlap after cortex based alignment was not uniform across all tested function areas, suggesting that some functional areas are strongly related to macro-anatomy e.g. the frontal eye fields (Paus, 1996) whereas others are not as tightly bound to gyri or sulci, e.g. the fusiform face area (FFA, but see Weiner & Grillspector (2010) and discussion in Frost & Goebel (2012)).
In order to overcome the problem that there remain differences in the spatial location of functional areas after registration to a standard template, and even after advanced alignment schemes, a region of interest (ROI) approach is often utilised. Restricting analysis to a few circumscribed, independently localised regions allows to carry out group comparisons of functionally corresponding regions across subjects with high statistical power and substantially reduces the correction needed for multiple comparisons as is needed for the many thousands of voxels or vertices of a whole brain (cortex) analysis. However this approach ignores activity in areas outside the ROIs which would be revealed by a whole-brain analysis. While one could perform both kinds of analyses separately (as is indeed reported in many published studies), an integrated approach combining ROI and whole brain analysis with maximal correspondence across subjects would be desirable. Here we propose an integrated alignment technique that incorporates the spatial specificity and statistical power of a region of interest analysis, with the benefits of a whole brain analysis (Frost & Goebel, 2013). To indicate that the whole-cortex alignment incorporates functional information from localised ROIs, we refer to the developed procedure as functionally informed cortex based alignment (fCBA). Due to the fact that a purely macro-anatomical alignment is not capable of successfully bringing functional regions fully into spatial correspondence, fCBA integrates both curvature (anatomical information) and functionally defined regions to produce a group brain, which is well aligned in terms of anatomy and function. The fCBA approach is, however, not limited to functional data and can incorporate any additional information such as subject-specific anatomical landmarks.
In order to apply the general gradient-based alignment procedure to POIs (defined as a contigous region of vertices in a cortex mesh), the functionally labelled areas are converted to a surface map and smoothed with a Gaussian kernel (approximated by iterative neighborhood smoothing) providing a gradient into the area’s neighbourhood. The gradient provides information to be followed during the alignment procedure maximising overlap across subjects. The integrated alignment process follows the same stages of standard group-CBA including a coarse-to-fine alignment of macroanatomical curvature information to a dynamic group average. The (smoothed) POI information is simply added as another constraint operating simultaneously with the curvature maps. The relative strengths of macro-anatomical (curvature-based) vs functional (smoothed POI based) forces is controlled by relative weighting parameters (see below).
Application
The execution of functionally informed CBA proceeds largely identical to a standard CBA process since fCBA merely adds additional constraints to the standard cortex-based alignment routine. Adding functional constraints to a CBA task is specified in the Cortex-Based Group Alignment Parameters dialog (see screenshot below) that can be invoked by clicking the Options button in the Align Group tab of the main Cortex-Based Alignment dialog. Below the CBA parameters table described in the Group Alignment topic, a section is available for specifying parameters for additional alignment itargets that is initially disabled (greyed out).
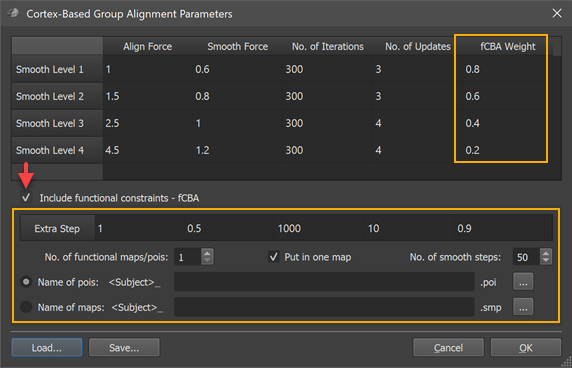
To turn on fCBA, check the Include functional constraints - fCBA option (see red arrow above). This will enable the options below that are specific for fCBA. Furthermore a new column, "fCBA Weight" is added in the general CBA parameters table in the upper section of the dialog (see orange rectangles above). The weights in the added column specify how strong provided functional information (weight wF) should drive the alignment relative to macro-anatomical (curvature) information (weight wA) with the constraint: wA + wF = 1. A value of "1.0" for the functional weight indicates that only functional information drives the alignment whereas a value of "0.0" indicates that only macro-anatomical information is used. The default values start with modest functional influence at the beginning of the CBA routine (0.2 for smooth level 4) and increase during the coarse-to-fine alignment scheme (0.4 for smooth level 3, 0.6 for smooth level 2 and 0.8 for smooth level 1).
In addition to functional forces during the 4 major stages of CBA, an additional 5th alignment stage is added that continues from the point when the most fine-grained alignment stage (smooth level 1) has been completed. The parameters of this additional stage can be specified in the single-row Extra-fCBA table that is located below the Include functional alignment option (see screenshot above). The reason for adding this step is to allow extra strong effects of functional constraints, i.e. if one wants to first run a rather standard macro-anatomical CBA and to add the fCBA step afterwards or in order to force alignment of corresponding functionally defined regions tolerating some macro-anatomical misalignment. As has been turned out (Frost & Goebel, 2013), the balance of forces during CBA combined with the possibility of an extra step provides a rather fine-grained control to achieve desired results (see below).
Besides setting parameters that control functional alignment forces, the corresponding regions that should be aligned need to be specified in subject-specific space. This is done by providing a POI file for each subject containing the definition of one or more corresponding areas. If more than one area is defined, the different regions need to appear in the same order in the POI file. If, for example, a localizer run has been performed to map a face area (pFus) and a place area (CoS, parahippocampal place area, PPA), a single .poi file with the pFus and the CoS POI needs to be created per subject and stored in the same order. The screenshot below shows part of the Patches-Of-Interest Analysis dialog showing the two POIs contained in the loaded POI file of the first subject (see orange rectangles).
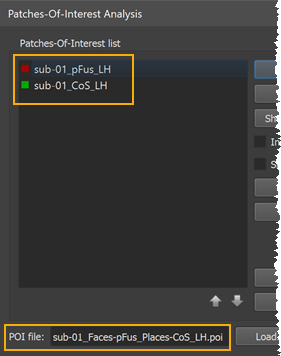
POIs can be created in the usual way, e.g. by running a MTC-GLM on individual SPH LH/RH cortex meshes followed by overlaying the desired contrast (e.g. "faces vs places"); after adjusting the threshold of the contrast t map appropriately, a target region (e.g. pFus) needs to be selected on the map as a contigous region by performing a CTRL-click (CMD-click on Mac); the marked region can then be converted into a Patch-Of-Interest (POI) by using the context menu entry Define Patch-Of-Interest (POI); if the order of the defined areas is not the same for each subject, the Up and Down arrow buttons in the Patch-Of-Interest Analysis dialog can be used to rearrange the POIs in a consistent order. The created subject-specific POI files need to be stored in the same folder that contains also the macro-anatomical alignment information, i.e. the curvature maps. Furthermore, the names of the POI files need to be identical except for the intial substring that must contain the subject ID - the same ID that is also used for the curvature maps. If, for example, a subject's curvature map has the name "sub-03_LH_SPH_CURVATURE.smp", the poi file need to also start with the "sub-03_" substring.
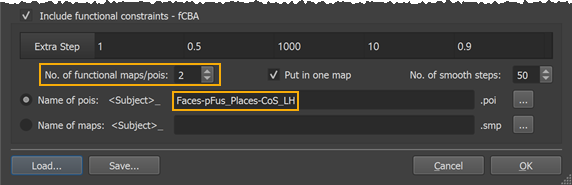
In the used example dataset, two pois (pFus, CoS) were defined using appropriate localizer runs and the .poi files for five of the subjects included in the example were named "[subID]_Faces-pFus_Places-CoS_LH.poi" (with subID = "sub-01", "sub-02", .. "sub-05"). In order to tell the program that these are the .poi names of the included subjects, only the file name sub-string following the subject-ID needs to be entered in the Name of pois text field without file extension (see lower rectangle in the screenshot above); with the entered POI file name template, the program derives the full POI file names for the individual subjects by prepending the subject-ID extracted from the curvature maps and by appending the ".poi" file extension. Furthermore, the number of POIs defined in each POI file needs to be set in the No. of functional maps/pois spin box (see upper rectangle in the screenshot above). Since in our example there are 2 POIs, the value entered is "2". In case the Put in one map option is turned on (default) one surface map file will be created per containing all smoothed POIs (POI maps) in a single surface map; this is achieved by forming a "max" operation on all individual POI maps. If this option is turned off, a separate surface map per POI is created allowing more fine-grained control over fCBA, which may be useful when areas are closely spaced. In that case, the table will show parameters for each separate surface map in the order (from left to right) as defined in the original POI/SMP files, e.g. the column "fCBA 1 Weights" contain the parameters for each curvature smoothing level for the first POI/SMP, the column "fCBA 2 Weights" for the second POI and so on. There are also as many columns added to the Extra Step table allowing each POI to have different force values during the 4 coarse-to-fine curvature alignment steps as well as during the extra-alignment step (using always the highest curvature resolution, i.e. leaset smoothed maps). Finally the No. of smooth steps value (for explanation of its function see below) can be set but it is recommended to keep the default value of "50". After all settings have been made, the Cortex-Based Group Alignment Parameters dialog can be closed by clicking the OK button.
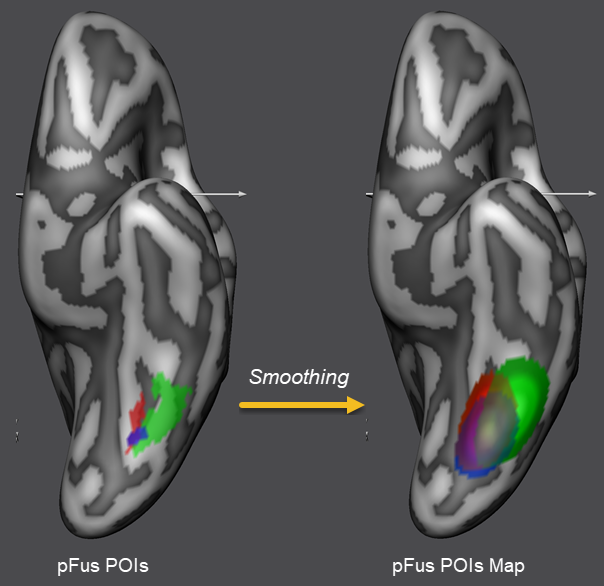
When now a dynamic group average CBA process is started (as usual by specification of the curvature maps of the corresponding hemispheres for the included subjects), the defined extra alignment constraints will be effective during alignment. Since the alignment process needs gradient information to work successfully, the POIs are first spatially smoothed before CBA begins so that they are represented with a peak value in the middle that gradually falls off if one moves away from the peak in any direction. In order to perform this smoothing operation, POIs are first converted into surface maps internally and saved to disk. In the snapshot above, the smoothed places area is shown for the included subjects using different colors. The amount of iterative neighborhood smoothing is specified by the No. of smooth steps value (see above). The performed smoothing spreads the binary region occupied by the original POIs into the neighborhood increasing the chance that corresponding regions across subjects (e.g. the faces and places POIs of all subjects) will overlap; note that functional alignment forces only have an effect if corresponding smoothed POIs overlap across subjects.
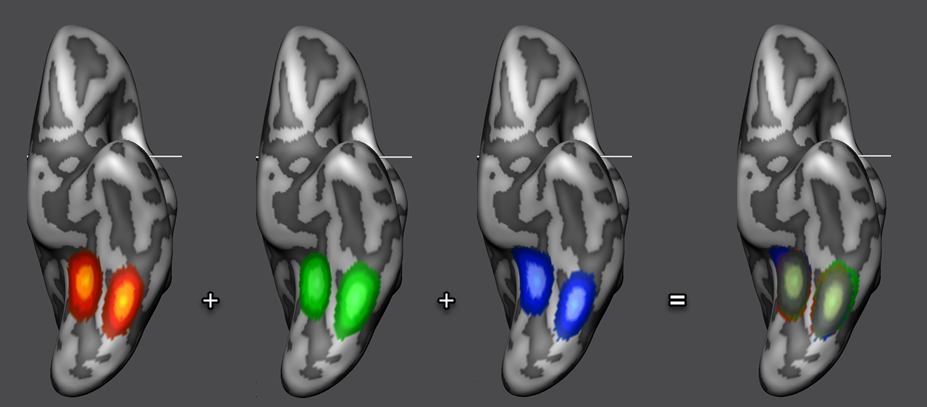
After fCBA has been completed, one can inspect the achieved POI (and macroa-anatomical) alignment in various ways. First, the program automatically puts the smoothed POIs in a final surface map that is displayed at the end of fCBA superimposed on a standard sphere or directly on the averaged folded group brain (if the individual "RECOSM_SPH" meshes can be located in the subject-specific sub-folders). The surface map is also stored to disk automatically and contains 4 macro-anatomical and 1 POI sub-map for each of the included subjects. In the overlaid surface map, only aligned curvature sub-maps are selected as default. In order to visualize the POI sub-maps, they need to be selected in the sub-maps list. Note that in order to visualize multiple maps simultaeneously, the Multiple selections option in the Advanced tab of the Surface Maps dialog needs to be turned on. In the snapshot above (right side), the POI sub-maps have been selected for each of the three subjects showing the nearly perfect alignment of the smoothed faces and houses POIs after fCBA. To visualize the different subjects better, the default orange sub-map color has been changed to a red, green and blue color value and the maps are shown also separately for each subject.
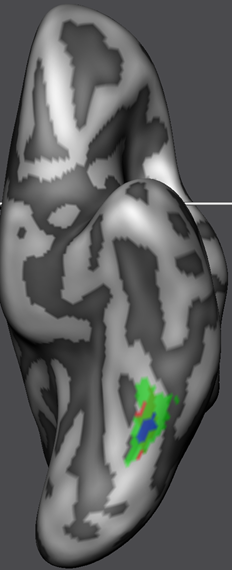
While the surface maps provide already insight in the quality of POI alignment, the alignment of the original POI shapes should be inspected by aligning the POIs using the standard CBA POI transformation tool. The snapshot above shows the resulting "ALIGNED" POIs on the right side and the location of the POIs prior to fCBA alignment on the left side (both are shown on the average folded group brain for comparison). As is evident, the POIs are indeed located at the same region after fCBA.
The fCBA procedure offers a powerful means to align corresponding functional regions across subjects. While the default functional weighting values provide good results, it may be necessary to adjust the values for specific purposes. While functional constraints should align corresponding POIs as good as possible, it is also desirable to achieve a good as possible macro-anatomical (curvature-based) alignment. The provided default values are set to optimize POI overlap tolerating visible decrease of macro-anatomical alignment as compared to running pure macro-anatomical CBA. Recently, optimal functional constraint parameters have been reported that appear well suited to integrate both goals in a near-optimal way (Frost & Goebel, 2013). While not always leading to a perfect POI alignment (i.e. POIs do not exhibit complete overlap after fCBA), parameters are suggested that lead to near-optimal POI alignment with minimal macro-anatomical detoriations as compared to pure macro-anatomical CBA. The functional weights for this "balanced" weighting for the four coarse-to-fine stages were 0.2, 0.4, 0.6, 0.9 with 400, 300, 300 and 1000 iterations with 4, 4, 3 and 10 number of updates, respectively. The recommended parameters for the extra step were 0.2 functional weighting, 400 iterations and 4 updates. While the provided default values and the values suggested here are good candidates to either push POI alignment or to go for a more balanced functional-macro-anatomical alignment, it should be noted that some experimentation with the parameters might be needed, i.e. by running fCBA repeatedly with different parameter sets.
References
Frost, M. & Goebel, R. (2013). Functionally informed cortex based alignment: an integrated approach for whole-cortex macro-anatomical and ROI-based functional alignment. NeuroImage, 83, 1002-1010.
Copyright © 2023 Rainer Goebel. All rights reserved.