BrainVoyager v23.0
Overview of EEG/MEG Distributed Source Analysis
Introduction
Modern high-resolution electroencephalography (EEG) and magnetoencephalography (MEG) techniques allow investigating non-invasively in humans spatially selective effects in electric and magnetic brain activity. When restricted to the channel (electrode or sensor) level, the spatial analysis of the EEG and MEG sources can only provide limited information about the cortical regions involved in the generation and the perturbation of these effects. Both wideband signals like event-related potentials (ERP) and event-related fields (ERF) and narrow-band (i.e. rhythmic synchronization) effects like event-related spectral perturbation (ERSP) and phase coherence (PC) can be originated from one or multiple cortical sites, thereby making the characterization of their cortical distribution potentially important for strengthening notions of functional specialization and building new augmented spatio-temporal models of functional integration.
In order to spatially characterize the cortical EEG and MEG sources with no or weak a priori assumptions about their location of origin, a distributed source imaging approach has been implemented and made available as a new module, called the EMEG Suite (see below) since BrainVoyager QX 2.0. The EMEG Suite has been updated to be compatible with BrainVoyager 21.2 and later.
Background
EEG/MEG channel time-series can be projected from the measurement extra-cranial space (the "channel" space) to a suitably prepared intra-cranial space (the "source" space) by placing an equivalent current dipole (ECD) in each node (vertex) of a surface mesh and estimating a distributed solution for the EEG/MEG inverse problem "constrained" to these dipole positions. Optionally, the orientation of the dipoles can also be constrained.
In order to illustrate the technical background of EEG/MEG distributed source modeling, let us consider a mesh with N vertices and a linear discrete ECD model as a data model. According to this model, M channel time-series representing the measurement m(t) can be expressed as linear combinations of N dipolar source time-series s(t):
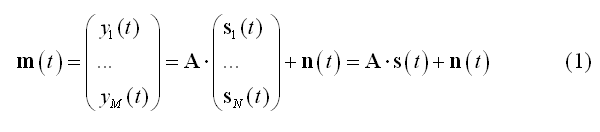
In (1) the columns of matrix A (M x 3*N) contain the lead fields of the dipolar sources for the given M-channel EEG/MEG configuration. si(t) = [sx(t), sy(t), sz(t)]t represents the source activity time-series of the current dipole placed on the i-th node of the mesh and n(t) represents the channel noise. The lead fields for the EEG/MEG configuration are computed according to a forward model of EEG and MEG activities, expressed by all possible distributions of the electric and magnetic fields outside the cranium due to unit current dipoles inside the brain. To reconstruct (and localize) the current sources underlying the signals in m(t), a linear inverse operator W needs to be estimated and applied to the measured signals:
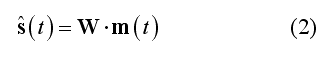
If N >> M, the linear problem in (1) is severely ill-posed and any reasonable (and unique) solution for W can only be achieved by posing mathematical and physical constrains on the output time-series (regularization). According to (2), all linear inverse distributed solutions to the EEG/MEG source reconstruction problem can also be seen as a collection of spatial filters (one per source dipole component and source dipole position defined across all channels) which can be subsequently applied to channel data for generating a source time-course at each and any vertex of the mesh. Matrix W (3*N x M) contains the spatial filter weights and s(t) is the estimated source time-series for all orientations and locations. Once the filter weights have been estimated (see Inverse Modeling), (2) can be used for reconstructing a point sourcetime-series at each vertex of the mesh (also called virtual intra-cranial electrode, see Source Reconstruction).
The EMEG Suite
How is it possible to perform the EEG/MEG distributed source modeling and analysis in BrainVoyager? A suite of functions and tools (called "EMEG Suite") is available, which allows a complete cortically constrained distributed imaging and analysis of EEG and MEG source activity. Starting from registered channel configurations and continuous data sets, you will end up with surface mesh time-courses (MTC) and maps (SMP) characterizing at the cortical source level your EEG and MEG signals. The newly generated EEG/MEG time-courses and maps will be different from typical fMRI time-courses and maps in their spatial and temporal physical resolution, but not in their format, thereby allowing a fully integrated EEG-fMRI and MEG-fMRI modeling and analysis. If your license supports the EMEG module, the EEG-MEG menu is available in the menu bar containing the Distributed Source Analysis item. The EMEG Suite and all other EEG-MEG tools address the following main points:
Forward modeling of the EEG/MEG sources for a given source space and a registered channel configuration. This step allows preparing the set of lead fields that are needed for inverse modeling. The lead fields are handled (and can be imported) as standard BrainVoyager collections of surface maps (SMP files) that must fit the selected source space (i.e. the surface mesh).
Channel data preparation and preprocessing. Continuous EEG/MEG channel time-course (CTC) data are combined with trigger information stored in protocol files (PRT) for epoching, averaging and time-frequency transformations. In this step EEG/MEG data sets are prepared for subsequent stages in the form of single-trial data (STD), average channel time-courses (ACT) and time-frequency data (TFD). During preparation, all extracted epochs (single trials) are scanned for artifacts and selectively rejected before averaging and time-frequency transformations. At this stage, all created ACT, STD and TFD can be plotted.
Covariance calculations on channel single-trial data sets (essential for inverse solution regularization).
Continous wavelet transform (CWT) filtering (applied at this or later stage of the analysis to obtain single-trial data with narrow-band complex signals). CWT filtering allows flexibly controlling the center frequency of the filter and the desired temporal and frequency resolution of the transform.
Inverse modeling. In this step the lead fields are combined with a channel covariance matrix for inverse model estimation. The inverse solution is, again, obtained as a collection of maps, (the inverse spatial filters) and stored as a new VMP or SMP depending on the source space. Minimum-norm and beamformers ara available as inverse approaches with possibility to further regularize the solution with typical mathematical constrains (e.g. depth-weighitng, LORETA, LAURA) and to incorporate spatial information about sources directly from fMRI activity via fMRI weighting.
Source time course reconstruction. In this step, an inverse solution generated in previous step can be combined with prepared averaged (ACT) and single-trial (STD) channel data sets, yielding the reconstructed source activity time-course at each position (vertex) of the source space (mesh). The source point is also called generically a virtual intra-cranial electrode. Source activity time-courses are stored in mesh time-courses files (MTC).
Source imaging and statistical analysis. This step allows combining multiple previously estimated inverse solutions (SMP) and prepared EEG/MEG channel data sets (ACT, STD, TFD) from multiple experimental conditions, sessions, subjects and groups in one common-space group-level distributed source analysis. Depending on the mode of operation, it is possible to produce group-level EEG/MEG source activity plots from predefined regions of interest (POI) or source images (maps) representing the activity in a defined time (or time-frequency) interval of interest. This is also a convenient tool for assembling and projecting data from individual channel configurations onto source spaces in batch, and later on summarize the results according to conditions, subjects and groups using other BrainVoyager tools, such as those for combining surface maps in simple and factorial statistical analysis (e. g. ANCOVA).
Source-level EEG-fMRI coupling effects in simultaneous EEG-fMRI experiments can be studied within the same source analysis framework (in source plot mode) and EEG-constrained time-course models (SDM) for fMRI can be obtained from a predefined regional source (POI).
Other EEG-MEG Tools and EEG-MEG Plugin
Before starting the EMEG Suite, EEG and MEG channel data, protocol information and electrode/sensor configurations should be made available according to the EEG/MEG file format and data specifications of BrainVoyager.
EEG and MEG raw data can be imported via the EEG-MEG Raw Data Import dialog and, when necessary, filtered via the EEG-MEG Data Filter dialog, both accessible from the EEG-MEG menu.
There are also EEG-MEG plugins accessible from the Plugins menu (EEG-MEG Plugins). For instance, anEEG/MEG Temporal independent component analysis (ICA) decomposition can be quickly obtained via the EMEG Temporal ICA (GUI) plugin. When EEG data are acquired simultaneously with fMRI, one may require to deal with typical fMRI artifacts induced on the EEG traces, such as the gradient artifact and the cardio-ballistogram artifact. If not already removed "on-line" during acquisition, two more computational plugins can be used (alone or in combination with temporal ICA) to remove these artifacts from EEG traces (see section on Simultanoues EEG-fMRI: FMRI Artifact Detection and Removal).
Finally, the EEG-MEG menu also offers the necessary surface tools to perform a maximally precise head-meash coregistration. This is essential for a correct distributed source EEG or MEG analysis, because the position of the channel must be made known by the program with respect to the position of the sources. To use these tools, the skin surface of the subject' s head should be reconstructed from a whole-head 3D T1 MRI and a set of at least three surface points (and the EEG electrodes) should be digitized to the same head. For more information about how the coordinates of the surface points, the EEG electrodes and the MEG sensors are handled in BrainVoyager, please consult the chapter on the EEG and MEG channel configurations.
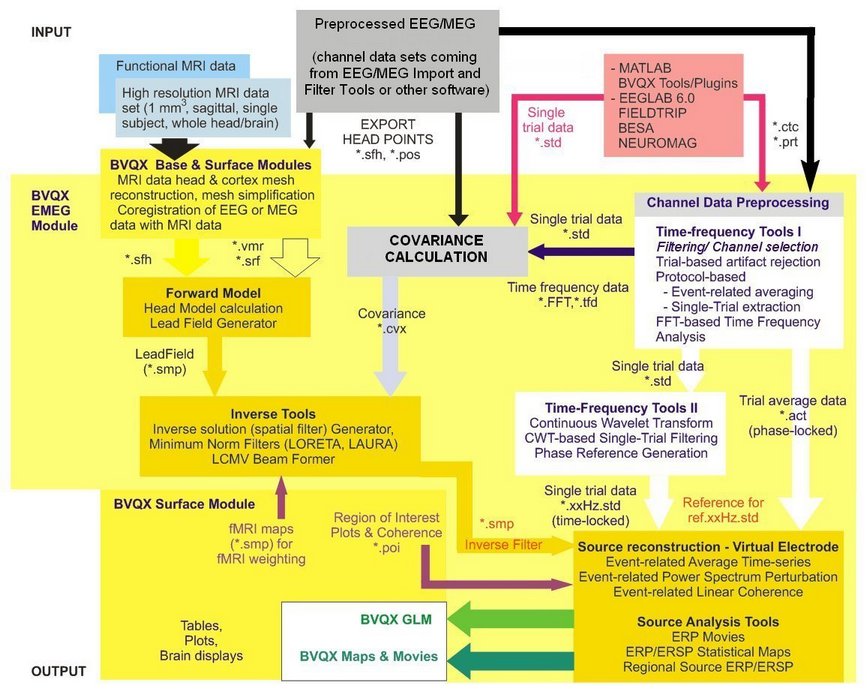
Below you find a schematic diagram of all functionalities of the EMEG Suite and their integration with other BrainVoyager tools and functionalities.
Copyright © 2023 Fabrizio Esposito and Rainer Goebel. All rights reserved.