BrainVoyager v23.0
FA Using Gradient-Driven Affine Transformations
A header-based initial alignment would produce a perfect match between the functional and 3D anatomical data sets if there would be no head movements between the two scans (and no other problems, such as EPI distortions). In order to correct for potential head motions, or more generally for any kind of suboptimal initial alignment, BrainVoyager QX attempts to further improve the coregistration using an intensity-driven alignment step after the initial alignment step. This subsequent alignment step can be applied after any of the initial alignment steps, i.e. after header-based alignment, corresponding points alignment or even after a simple sagittal reorientation of the functional data. The only requirement of the fine-tuning alignment step is that both data sets are in BrainVoyager's standard sagittal orientation.
Automatic coregistration algorithms often use the sum of squared intensity differences between the source and target data set as a "goodness-of-alignment" measure evaluating the quality of alignment, i.e. parameters (e.g. translations and rotations) are to be determined in such a way that the goodness-of-alignment measure reaches a minimum (usually in an iterative procedure). Unfortunately the squared intensity difference measure (used in the second available automatic FA procedure) may not always lead to the optimal coregistration state since it is not robust in case of (non-removed) spatial inhomogeneities. The more powerful and robust FA procedure described here does not use intensity per se but gradient information as its goodness-of-alignment measure. While intensity differences strongly depend on inhomogeneities in the data, gradient information is largely unaffected since it evaluates the change of signal intensity with respect to neighboring voxels. More specifically, the implemented "Normalized Gradient Field" (NGF) algorithm considers a source and target volume as aligned, when intensity changes in the same way in both volumes at corresponding positions. Another advantage of this alignment routine as compared to the intensity-driven alignment method is the (optional) inclusion of scale and shear parameters extending the performed 6 parameter rigid (3 translations + 3 rotations) alignment to 9 parameter (rigid + scale) and full affine (12 parameter) transformations. As default, the program starts with a rigid alignment followed by a 9 parameter alignment. The included fit of 3 scale parameters seem to be especially beneficial for some EPI images with different "stretching" in image space along the frequency and phase encoding gradient. The FMR-VMR Alignment Options dialog available in the Fine-Tuning Alignment tab of the FMR-VMR Coregistration dialog allows to include also the full 12 parameter alignment when desired. When turned on, a pre-alignment step will search for good starting parameters; this initial explorative search step can be often even used in case that the FMR and VMR files were not scanned in the same session. The achieved coregistration will be visualized using the the edge overlay display option for the fused source and target data sets.
After the initial alignment step has been performed, you can start the gradient-driven alignment procedure. Invoke the FMR-VMR Coregistration dialog by clicking the Align button in the FMR-VMR coregistration field of the Coregistration tab of the 3D Volume Tools dialog. Switch to the Fine-Tuning Alignment tab. Since gradient-based alignment is the default mode for the fine-tuning coregistration step, the respective Gradient-based affine alignment (6 - 12 parameters) option will be checked as default. If this is not the case, click this option to select this fine-tuning alignment method (see snapshot below).
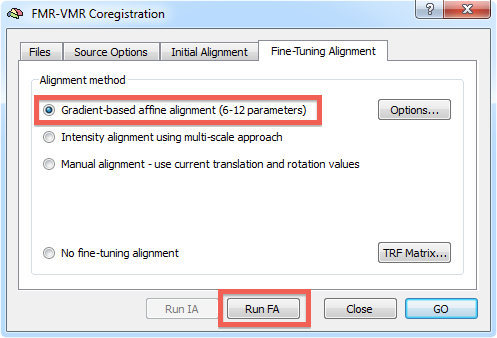
To start the alignment, click the Run FA (or GO) button. During the alignment procedure, information printed to the Log pane informs about the progress made. You can observe the iteratively updated parameter estimates during alignment in the Coregistration tab of the 3D Volume Tools dialog. The snapshot below (bottom part) shows the estimated parameters in the Translation and Rotation field in the 3D Volume Tools dialog after the fine-tuning adjustment has been completed. The lower row of the VMR window shows the functional and anatomical volume in edge blend mode. Details of different visualization options for the source and target volume can be found in the IA using header-based coregistration topic.
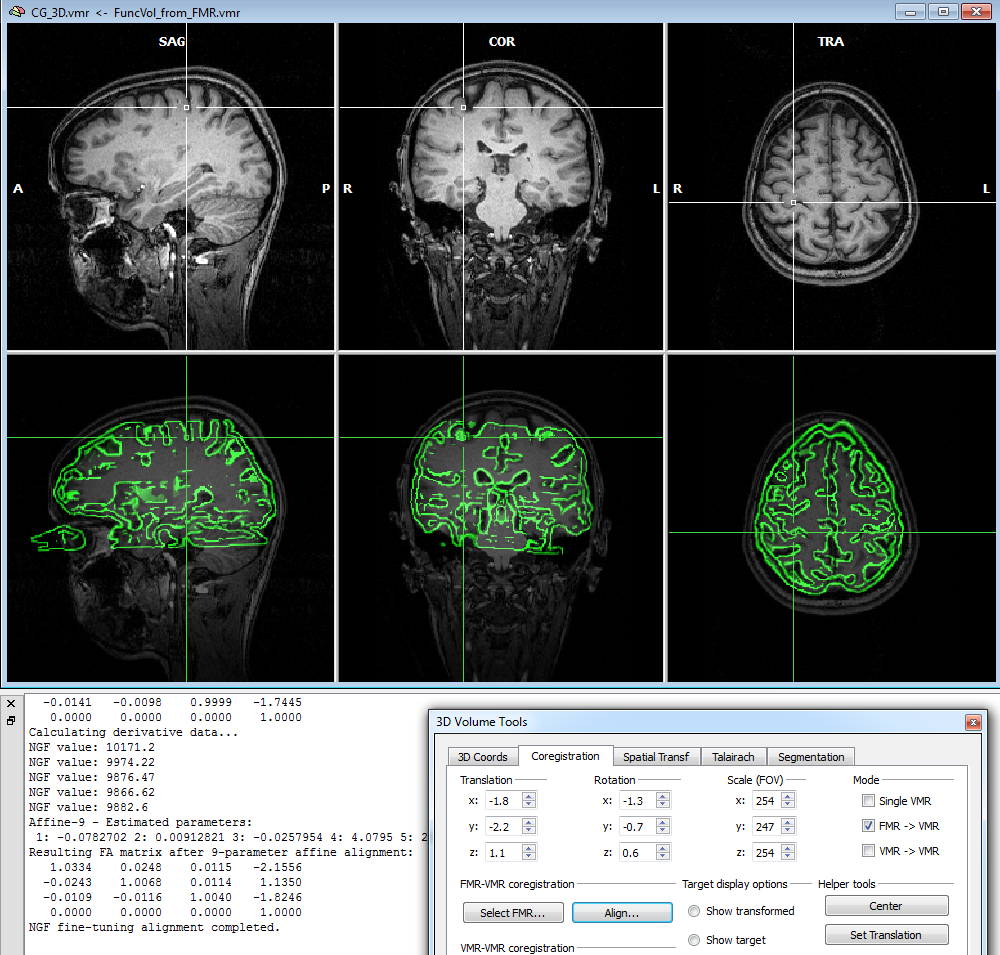
Note that in the displayed example, the VMR data set was used prior to running intensity-inhomogeneity correction (IIHC) in order to reveal that this FA method is robust against spatial inhomogeneities. It is, however, recommended to run IIHC as the first step after creation of a new VMR file since this will improve many subsequent anatomical processing steps, including automatic Talairach transformation and cortex segmentation.
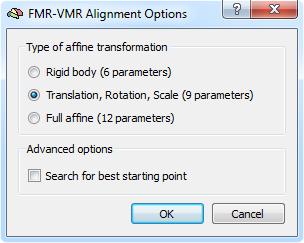
The gradient-based alignment fits as default 9 parameters (3 translations, 3 rotations and 3 scales). In case that there is evidence for shear transformations between the EPI volume and the anatomical volume, the default model can be extended to a full 12 affine parameter model. In order to switch to the 12 parameter model, click the Options button in the Fine-Tuning Alignment tab of theFMR-VMR (or DMR-VMR) Coregistration dialog that will invoke the FMR-VMR Alignment Options dialog (see snapshot above). Besides the Full affine (12 parameters) option and the default 9 parameter model, you may also select the simpler Rigid body (6 parameters) model.
Copyright © 2023 Rainer Goebel. All rights reserved.