BrainVoyager v23.0
Coregistration of Functional and Anatomical Data Sets
The coregistration routines in BrainVoyager usually enable a fully automatic, highly precise alignment of functional (FMR) and anatomical (VMR) data sets. To guarantee a good alignment result in any situation, the coregistration process has been split into two main stages:
The goal of the initial alignment step is to bring the functional and anatomical data sets in close proximity from a potentially very different starting point. If, for example, the functional data is recorded in a different field-of-view and different slice orientation than the anatomical data, the intial alignment should bring them at least into overlapping space with the same global orientation. This alignment step usually requires large rotation and translation values. The fine-tuning alignment step on the other hand assumes that the two data sets are already pretty close to each other, which may allow iterative minimization techniques to do an automatic fine adjustment of the alignment in a similar way as during 3D motion correction. To allow a successful coregistration in any situation, both the initial and fine-tuning alignment step can be performed in several ways. In the optimal situation, the functional and anatomical data sets are from the same scanning session and information about the scanning parameters (slice positioning) is available for both datasets, e.g. from the original DICOM files or corresponding NIfTi files. In this situation, the initial alignment performs a mathematically exact coregistration of the functional and anatomical data. Since the subject, however, might have made small head movements between the functional and anatomical same-session scans, small additional adjustments might be necessary, which can be performed using the fine-tuning alignment step.
There are two methods available for the fine-tuning alignment step, a gradient-based intensity-driven alignment, and (since BrainVoyager 20.2) boundary-based registration (BBR). The gradient-based method is simpler to apply but in challenging cases, the BBR method produces better (more robust) results. BBR requires, however, the creation of a cortex mesh, which is not necessary in case of intensity-driven alignment.
Source and target. The coregistration routines do not change the space (coordinate system) of the anatomical (VMR) data serving as the target for the functional (FMR) data, which is scaled, translated and rotated to match the space of the VMR data set. The scaling operation of the functional data is performed as the first step by transforming a set of functional slices into a volume with the same resolution as the VMR file. For this scaling transformation, the program uses the voxel resolution values stored in the selected FMR file. Since both brain datasets originate from the same person, the alignment routines subsequently perform a rigid body transformation (rotations and translations). The mathematical alignment is in principle able to perform additional scaling and shearing adjustments (full affine trasformation) but these options are turned off in the present implementation. In the context of functional and anatomical coregistration, we refer to the FMR data set as the "source" and the VMR data set as the "target" of alignment.
Which VMR file? To start the alignment process, one needs to load the VMR file to which the functional data should be aligned. If you have, for example, recorded a 3D anatomical data set in the same session as the functional data, open the respective VMR file. In case your original 3D data was not in BrainVoyager's standard sagittal view and/or did not contain 1 mm isotropic voxels, it is advised not load the originally created VMR file but the one which has been transformed into the standard sagittal view with 1mm iso voxels. If appropriate header information was available during project creation, BrainVoyager will have automatically performed the necessary standardization steps and you should have a file with an extension such as "_ISO_SAG.vmr" (or "_ISO.vmr" or "_SAG.vmr", if only one of the two standardization steps were necessary). In many cases, the 3D anatomical scan might already been recorded with 1.0 mm resolution and with sagittal slices and in this case these transformation steps are not necessary. For details on standardization of original VMR data sets, see topic xx.
Most importantly, it is highly recommended to run the intensity inhomogenity correction procedure and to use the resulting "_IIHC.vmr" file for FMR-VMR coregistration. This is important for the fine-tuning adjustment step since the brain is extracted from head tissue and intensity inhomogeneities are reduced leading to more robust fine-tuning alignment results. In the sample data used in the snapshots below, the original intra-session VMR file has been created as "CG_3DT1MPR.vmr" from DICOM slices. Since the data was recorded in sagittal orientation with a 1 mm isovoxel resolution, no spatial transformations were performed by BrainVoyager. The data was, however, inhomogeneity corrected and therefore the file "CG_3DT1MPR_IIHC.vmr" is used as the target for coregistration.
If you have loaded the proper target VMR file, the 3D Volume Tools dialog will appear automatically. Enlarge the dialog by clicking the Full dialog >> button and then switch to the Coregistration tab. The alignment of functional and anatomical data sets is performed using the FMR-VMR Coregistration dialog, which can be called by clicking the Align button in the FMR-VMR coregistration field of the Coregistration tab (see snapshot below).
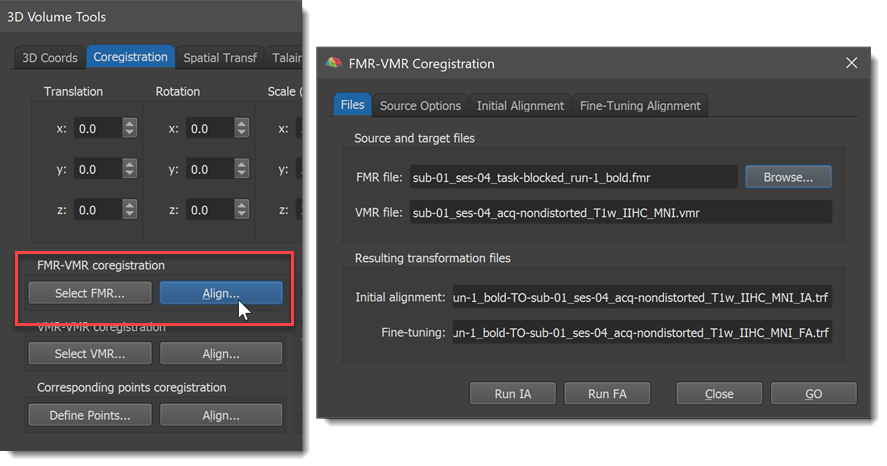
The VMR file: text box in the Source and target files field of the Files tab shows the currently loaded target VMR file, which is "CG_3DT1MPR_IIHC.vmr" in the example snapshot above. In order to align a functional data set to the loaded target VMR data set, you must first specify the respective FMR file using the Browse button in the Source and target files field (you might also choose the FMR source file prior to calling the FMR-VMR Coregistration dialog by using the Select FMR button in the FMR-VMR coregistration field of the Coregistration tab of the 3D Volume Tools dialog). After selection of the source FMR file, the Files tab of the FMR-VMR Coregistration dialog shows the selected source FMR file in the FMR file: text box as well as names for the transformation files, which will be produced and automatically saved when running the respective alignment steps. The transformation files are in the Initial alignment: and Fine-tuning: text boxes of the Resulting transformation files field.
In the example snapshot above, the source FMR file "CG_OBJECTS.fmr" has been selected. From the source FMR and target VMR files, the program generates the two resulting alignment file names by concatenating the FMR file name with the substring "-TO-" and the name of the VMR file. The two file names end with the substring "_IA.trf" and "_FA.trf" respectively.
In case one uses same-session data and know that both the functional and anatomical data contain header information about their location and orientation in scanner space (e.g. when created from DICOM files or when opening scanner-space NIfTI files), one can simply click the GO button to start the initial and fine-tuning alignment steps. If these conditions are not met or when one wants to usse BBR alignment, the two alignment steps should be launched separately, i.e. by clicking the Run IA button and checking the visual display assessing the result. If there are messages about missing header information, additional options are available that are described in the respective initial alignment and fine-tuning alignment help sections. In case the initial alignment was successful, the Run FA button can be clicked if one wants to use the gradient-based alignment approach. If one wants to use the BBR method, one should follow the instructions in section Boundary-Based Registration.
Which FMR file? In the example above, the FMR file, which was created from the raw data has been selected. As alternatives, one could have selected the FMR file of the first volume, i.e. "CG_OBJECTS_firstvol.fmr". In case that the true T1-saturated first volume of a functional scan is available (and not discarded already at the scanner site), the first volume is the optimal volume for the intensity-driven fine-tuning alignment step. If not turned off, BrainVoyager creates automatically also an AMR file of the first volume during project creation. For visualization purposes, this file is automatically attached to the standard FMR file, i.e. "CG_OBJECTS.fmr". This linked AMR file of the first volume may also be used for the intensity-driven fine-tuning alignment step. It is, however, recommended to select the FMR file but make sure to use the unpreprocessed file (e.g. "CG_OBJECTS.fmr") and not any preprocessed FMR (e.g. "CG_OBJECTS_3DMC_SCSAI_LTR_THP3c.fmr"). In case of multi-run data, it is even recommended to use the preprocessed file since the linked "_firstvol_as_anat.amr" file is replaced by a "_CoregFirstVol.amr" file for subsequent runs that are aligned to a reference (usually first) run (see also section "Implications For FMR-VMR Coregistration" in topic Motion Detection and Correction). Whether to use an FMR or AMR file can be specified in the Source options tab of the FMR-VMR Coregistration dialog (see screenshot below).
As described above, the default option in the Options for FMR/AMR source field is Use FMR data (EPI slices) opton. To use an AMR file instead, select the Use linked AMR option. Another option in this field is the Invert intensities option, which is turned on as default: By inverting the intensities of T2* weighted functional data, the resulting images look similar to T1 weighted anatomical data. This inversion is important for the intensity gradient driven adjustments during the fine-tuning alignment step. If you have linked a true anatomical coplanar set of slices (i.e. a special high-resolution T1 weighted scan of the functional slices), you should turn off this option. For information about the Correct inhomogeneities opton, go to topic Intensity Inhomogeneity Correction of Functional Volume.
The Flip slice order and the To SAG options in the Additional options for manual alignment field are only relevant if there is no header information available for the initial alignment step. In this case, it might be sometimes necessary to be able to flip the order of slices of the functional data, which can be achieved by turning on the Flip slice order option. If the initial alignment is done by the "corresponding points method", it is helpful to first orient the functional data into BrainVoyager's standard sagittal view, which can be done using the To SAG option. If this option is greyed out, click the Create volume button in the Options for FMR/VMR source field first.
Copyright © 2023 Rainer Goebel. All rights reserved.