BrainVoyager v23.0
Intensity inhomogeneity Correction of Functional Volume
When using some modern phased array coils for MRI scanning, intensity inhomogeneities are not only observed for anatomical data but also for functional data affecting visualization of statistical maps overlaid on functional data. More importantly, inhomogeneities in functional data may reduce the quality of intensity and gradient-driven fine-tuning alignment (FA). While BrainVoyager's main FA routines (BBR and gradient-driven alignment) are less affected than simple intensity-driven alignment, they might produce sub-optimal results in case of strong intensity inhomogeneities in the functional data. An important option (introduced in BrainVoyager 21.4) is thus the possibility to correct inhomogeneities in the created functional source volume used for fine-tunign alignment. If functional IIHC is enabled, the created functional volume after the initial alignment step will be used as input and the same intensity inhomogeneity correction (IIHC) procedure will be performed as used for anatomical data. The resulting corrected volume will be used as input for the subsequent fine-tuning (FA) alignment step. Note that the IIHC-corrected functional volume is automatically also transformed back into the FMR-STC slice space creating inhomogeneity corrected AMR files for visualization (see below).
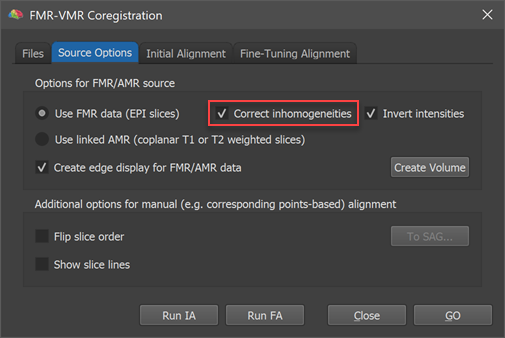
Running IIHC on Functional Volume for FA Alignment
As usual, the FMR-VMR alignment can be started after loading a native-space anatomical volume (VMR) file. The functional datset that one wants to align to the current VMR needs to be specified in the FMR-VMR dialog as usual. It is important to select a FMR file (not an AMR file) to enable the possibility of functional IIHC (AMR files only store 1-byte data while FMR-STC files store 4-byte float (or 2-byte int) data required for IIHC). The Correct inhoogeneities option is thus only enabled in case that the Use FMR data option is selected in the Options for FMR/AMR source in the Source Options tab (see screenshot above). As recommended, the selected FMR-STC dataset should not have been spatially smoothed in order to keep as much as possible anatomical detail, i.e. it is advised to select the originally created "[name].fmr" file or the "[name]_firstvol.fmr" file.
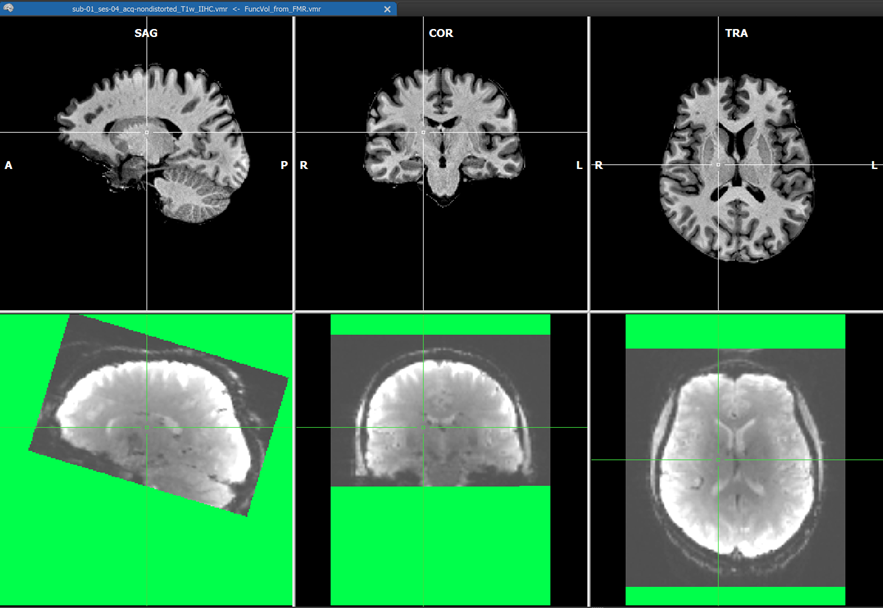
After (header-based) initial alignment (launched by clicking the Run IA button), the functional volume is brought roughly in the same space as the native-space anatomical data using the created IA transformation matrix. At this point, i.e. between IA and FA alignment, the intensity correction is performed on the IA-aligned functional volume if enabled. The screenshot above shows example data with substantial inhomogeneities, which is shown during the functional IIHC calculation (green color shows regions without functional data).
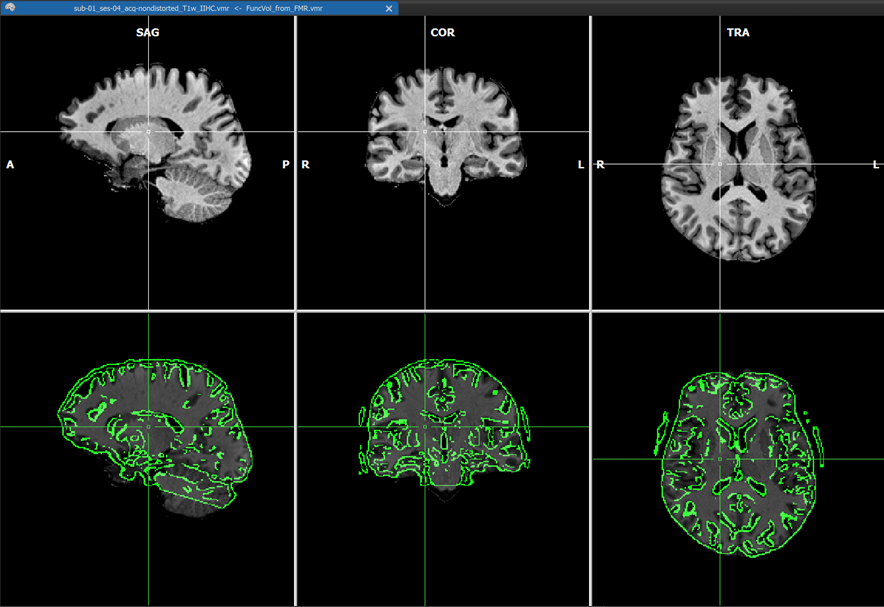
The coordinates of the non-background (brain) voxels in the IIHC-VMR are used to mark the respective voxels in the functional volume to define the space used to estimate the intensity inhomogeneities ("bias field") in the functional volume. Since the VMR is used to label voxels in the functional volume for IIHC, it is recommended to use a "skull-stripped" anatomical IIHC VMR is used. Note that after a standard anatomical IIHC preprocessing pipeline, the native-space VMR will not only be intensity inhomogeneity corrected but the brain will also be extracted as long as the Include mask generation (brain extraction) option is turned on (default) in the 16 Bit 3D Tools (IIHC) dialog; no extra brain extraction step prior to anatomical IIHC is thus required. If, as recommended, a VMR file is used as target that has been intensity inhomogeneity corrected, the Correct inhomogeneities option in the Options for FMR/AMR source field is turned on as default when selecting a source FMR file. Results should still be acceptable if a non-skull-stripped VMR is used as input but in that case the Correct inhomogeneities option is turned off and one needs to turn it on manually if one wants to apply functional IIHC. After the IA step - including functional IIHC - has completed, one can see the corrected volume (with inverted intensities) by usng the usual toggles in the Target display options field in the Coregistration tab of the 3D Volume Tools dialog (or by using the corresponding F keys). The visualizations during the IA step is slightly different than when not performing functional IIHC. The screenshot with the green mask shows the created and IA aligned functional volume before correction. The screenshot above shows the result after correction using the default "blend edges" quality check display with calculated edges (along gradients) of the IA coregistered functional volume superimposed on the target anatomy.
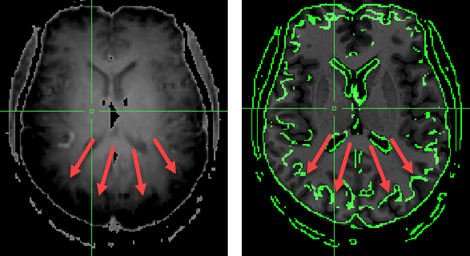
When functional IIHC is not performed, edges may be wrongly detected at regions where intensities change strongly (see screenshot above). The red arrows show where intensities rise strongly towards the posterior end of the brain (left panel, note that intensities are shown inverted). When subsequently running gradient-based FA using the non-corrected example data, the alignment will fail highlighting the importance of functional IIHC.
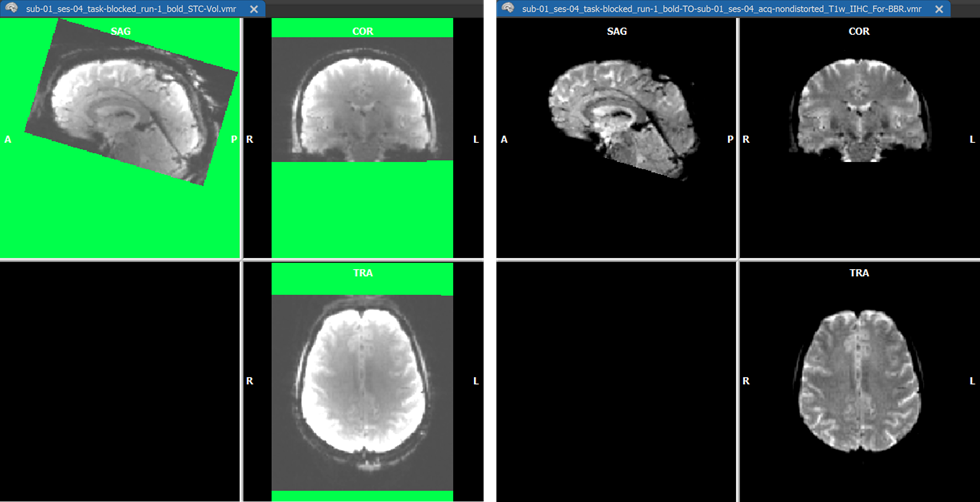
For visual comparison (and potential use), the functional volume is also stored to disk before and after IIHC both as a VMR and a V16 file (see screenshot above). The created functional volume before IIHC is saved under the name "[fmr-file]_STC-vol.vmr" and "[fmr-file]_STC-vol.v16". The corrected volume is also stored to disk under the usual name "[fmr-file]_TO_[vmr_file]_For-BBR.vmr" ready for use with BBR fine-tuning alignment. In addition, a.V16 version of the corrected volume is saved to disk as "[fmr-file]_TO_[vmr_file]_For-BBR.v16". With the corrected functional volume one can proceed as usual with the graident-based FA step or with BBR FA to complete the FMR-VMR coregistration.
Using IIHC Functional Volume for FMR Visualization
While the main purpose of the intensity inhomogeneity correction of the functional volume is for enhancing the quality of fine-tuning FMR-VMR alignment, the corrected volume can also be used to create an inhomogeneity corrected set of slices for visualization of the FMR data itself. For this purpose, the corrected volume is "back-projected" from the VMR space into the original slice space by inverting the IA spatial transformation matrix. The program creates an FMR file with the name "[file_name]_IIHC.fmr" that refers to the STC data file containing the corrected (single) volume: "[file_name]_IIHC.stc". Furthermore, the functional volume is converted into an AMR file for visualization purpuses ("[file_name]_IIHC.amr"). This file can now be used for visualization purposes by selecting it in the FMR Properties dialog for the original FMR file as well as for any preprocessed version. For the original FMR file, this is actually done automatically - when opening the file, the improved visualization AMR file will be shown instead of the original "[file_name]_firstvol_as_anat.amr" file. In case of strong inhomogeneities as in the example data used here, the inhonogeneity corrected AMR visualization file will substantially enhance the visualization of the slices, especially when overlaying functional maps (see below). Note that this process does not change the original functional data in any way, i.e. it is only used for enhanced visualization purposes.
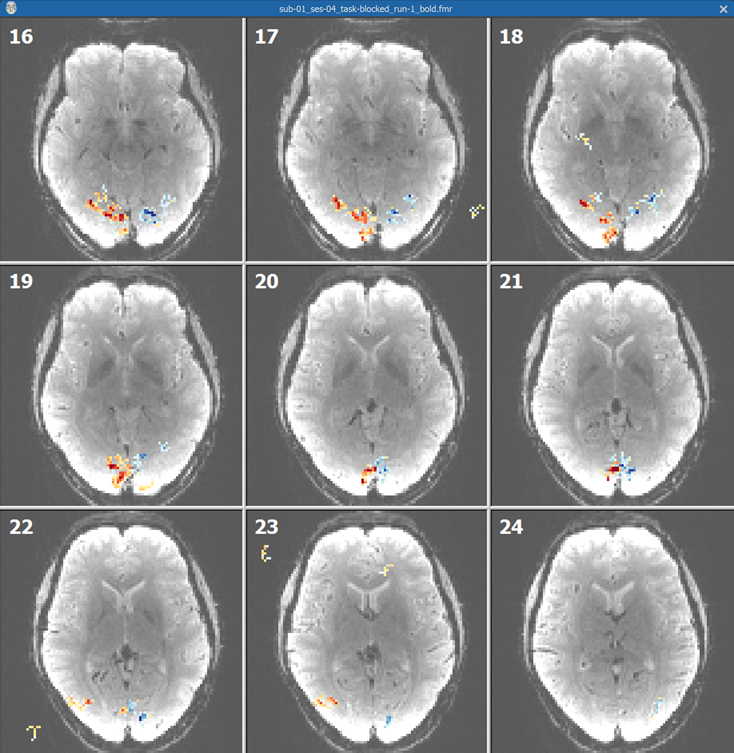
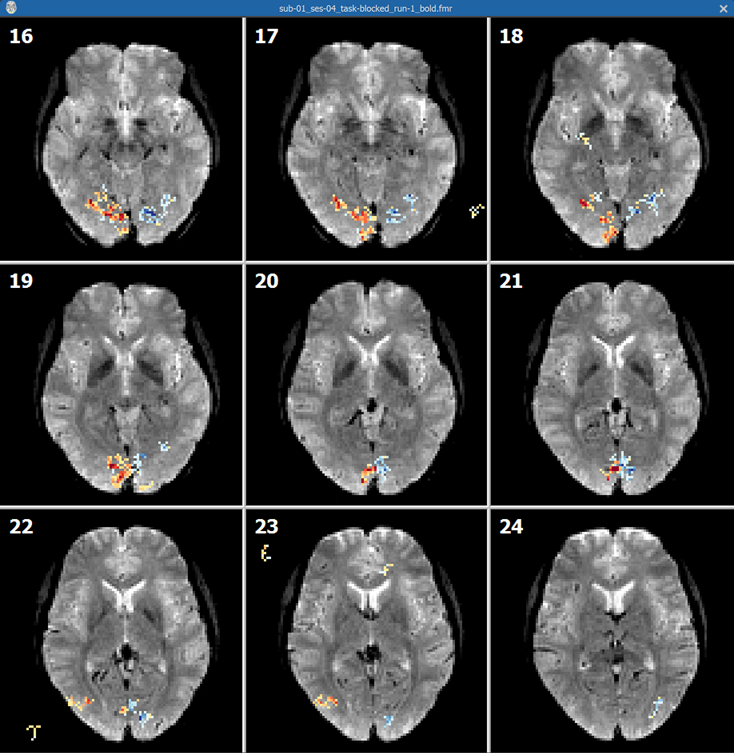
Ths screenshot above shows some slices from the unpreprocessed FMR document created from the "Getting Started Guide" dataset with an overlaid "left vs right visual field" contrast map. The slices clearly exhibit strong intensity inhomogeneities with the outer (cortical) regions of the brain much brighter than the medial brain regions. The screenshot below shows the same slices when using the inhomogeneity corrected slices exhibiting much less inhomogeneities aiding in relating functional activation to underlying anatomy.
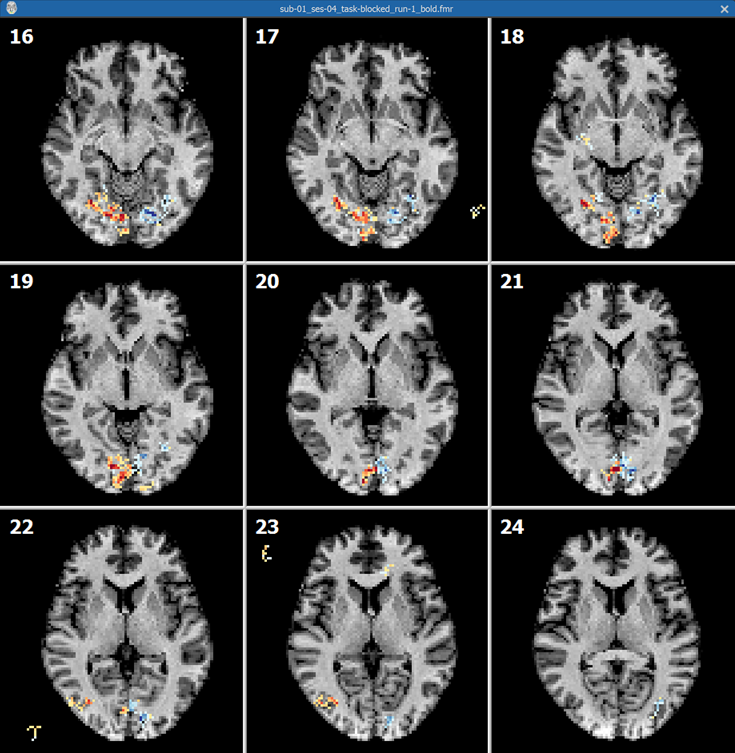
Besides the corrected functional volume, the program also back-projects the IA aligned VMR data back into the FMR slice space allowing to visualize functional activation on anatomical slices. The back-projected AMR visualization file is identified by the name "[file_name]_T1.amr". In the screenshot below, the T1 file has been selected in the FMR Properties dialog showing the functional map overlaid on the back-projected VMR data.
Copyright © 2023 Rainer Goebel. All rights reserved.