BrainVoyager v23.0
NIfTI Sidecar Files - JSON and TSV
When creating NIfTI files from original scanner (e.g. DICOM) data, BrainVoyager creates additional JSON and tab-separated value (TSV) sidecar files that fulfil two purposes:
- BIDS compatibility. As required by BIDS, the sidecar files store relevant information from the DICOM header in a human-readable form; in addition, information about task events for functional runs is stored in a TSV file in case a protocol is available.
- BrainVoyager-specific header information. BrainVoyager adds to the saved JSON file a “BrainVoyagerInfo” entry in order to store information that is specific to BrainVoyager, i.e. information that is usually stored in the header of FMR, DMR and VMR files and for which the NIfTI header alone is not sufficient.
More details about BIDS (Brain imaging Data Structure) is described in topic BIDS Compatibility in the Data Analysis Management chapter.
JSON Sidecar Files
When loading a NIfTI file, BrainVoyager checks whether a JSON file with the same name exists. If such a sidecar JSON file is found, it will be read and used to access additional metadata that is not stored in the NIfTI header. The JSON file complements the NIfTI header containing information about the data acquisition as specified in the BIDS format. In case that the NIfTI file was created by BrainVoyager using the Data Analysis Manager or the Create Document Wizard, the file will include also information that is specific to BrainVoyager and usually stored in the header of FMR and VMR files. A NIfTI file together with such a JSON sidecar file can thus completely replace a FMR-STC or VMR/V16 native file format. The BrainVoyager specific data is stored in the “BrainVoyagerInfo” entry, which is a JSON oject inside the root-level jSON object.
For 3D anatomical (VMR) documents (see example file below), a BrainVoyager created JSON file contains information about the BrainVoyager document type ("DocumentType"), the framing cube dimension in which the data is centered ("FramingCubeDim"), the reference space ("ReferenceSpace"), a list of eventually performed spatial transformations and preprocessing steps and values calculated from original (e.g. DICOM) data files such as the location of the center of the first and last slice in the original scanner coordinate system ("CalculatedDicomSlice1CenterX/Y/Z" and "CalculatedDicomSliceNCenterX/Y/Z" values). These are relevant values for BrainVoyager that are usually stored in the header section of VMR files (other header values, like the dimensions of the data are stored in the NIfTI header).
Example JSON 3D anatomical sidecar file "sub-01_ses-01_T1w.json":
{
"AcquisitionNumber": 1,
"AcquisitionTime": "154932.407494",
"BrainVoyagerInfo": {
"CalculatedDicomSlice1CenterX": -95.5,
"CalculatedDicomSlice1CenterY": -15.343368530273438,
"CalculatedDicomSlice1CenterZ": -30.53614044189453,
"CalculatedDicomSliceNCenterX": 95.5,
"CalculatedDicomSliceNCenterY": -15.343368530273438,
"CalculatedDicomSliceNCenterZ": -30.53614044189453,
"CoordinateSystem": 1,
"DocumentType": "VMR",
"FramingCubeDim": 256,
"NrOfPastSpatialTransformations": 0,
"NrOfPreprocessingSteps": 0,
"ReferenceSpace": "Native",
"Version": 1
},
"ConversionSoftware": "BrainVoyager",
"ConversionSoftwareVersion": "21.0.0.3592",
"ImageNumber": 192,
"ImageOrientationPatientDICOM": [
0,
1,
0,
0,
0,
-1
],
"ImagePositionPatientDICOM": [
95.5,
-142.84337,
96.963856
],
"ImageType": "ORIGINAL PRIMARY M ND",
"MagneticFieldStrength": 2.89362,
"Manufacturer": "SIEMENS",
"ManufacturersModelName": "Trio",
"Modality": "MR",
"PatientPosition": "HFS",
"RepetitionTime": 2300,
"SeriesDescription": "t1_mprage_sag_std",
"SeriesNumber": 2,
"SliceThickness": 1,
"StudyDescription": "RaiGoe^standaard"
}
For 4D functional (FMR-STC) documents (see example file below), a BrainVoyager created JSON file contains similar information as for VMR files (e.g. "DocumentType", "CalculatedDicomSlice1CenterX/Y/Z" and "CalculatedDicomSliceNCenterX/Y/Z" values) but it adds important entries with respect to the temporal acquisition of the data, including information about slice scanning order ("SliceAcquisitionOrder") and, if available in Siemens DICOMs, also a slice timing table that specifies when each slice of a volume was recorded relative to the first recorded slice(s); note that the slice scanning table also handles multiband (simultaneous multi-slice) acquisitions since more than one slice can have the same slice timing value. These (and other) values are relevant for BrainVoyager and usually stored in the header section of FMR files. In case that a prepared protocol file was available when the NIfTI file was saved to disk, it will be also embedded in the JSON sidecar file as a "Protocol" object inside the "BrainVoyagerInfo" object with information about the condition names, colors and intervals in the same way as usually stored in a PRT file.
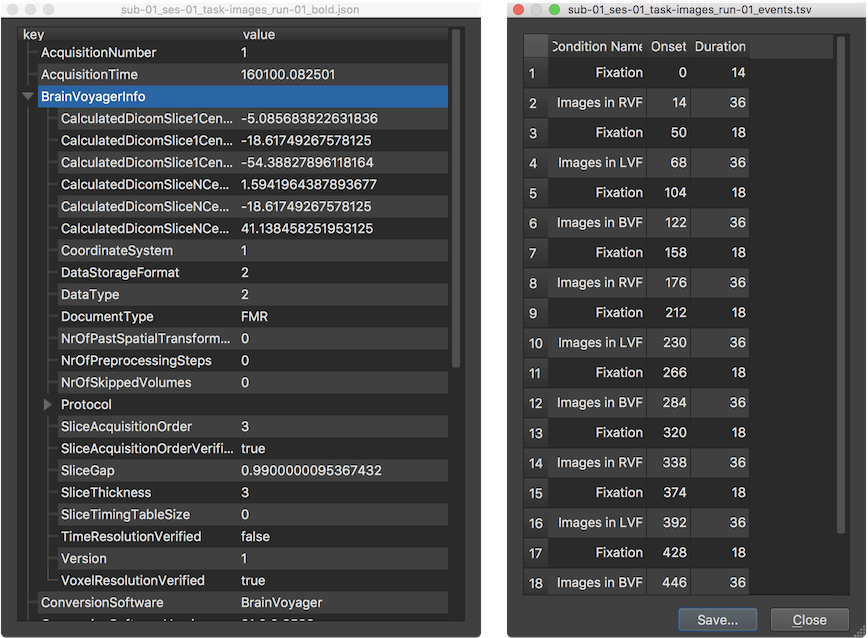
Example JSON functional sidecar file "sub-01_ses-01_task-images_run-01_bold.json":
{
"AcquisitionNumber": 1,
"AcquisitionTime": "160100.082501",
"BrainVoyagerInfo": {
"CalculatedDicomSlice1CenterX": -5.085683822631836,
"CalculatedDicomSlice1CenterY": -18.61749267578125,
"CalculatedDicomSlice1CenterZ": -54.38827896118164,
"CalculatedDicomSliceNCenterX": 1.5941964387893677,
"CalculatedDicomSliceNCenterY": -18.61749267578125,
"CalculatedDicomSliceNCenterZ": 41.138458251953125,
"CoordinateSystem": 1,
"DataStorageFormat": 2,
"DataType": 2,
"DocumentType": "FMR",
"NrOfPastSpatialTransformations": 0,
"NrOfPreprocessingSteps": 0,
"NrOfSkippedVolumes": 0,
"Protocol": {
"Conditions": [
{
"ColorB": 40,
"ColorG": 40,
"ColorR": 40,
"IntervalsFrom": [
1,
26,
53,
80,
107,
134,
161,
188,
215,
242
],
"IntervalsTo": [
7,
34,
61,
88,
115,
142,
169,
196,
223,
250
],
"Name": "Fixation",
"NrOfIntervals": 10
},
{
"ColorB": 77,
"ColorG": 84,
"ColorR": 217,
"IntervalsFrom": [
35,
116,
197
],
"IntervalsTo": [
52,
133,
214
],
"Name": "Images in LVF",
"NrOfIntervals": 3
},
{
"ColorB": 72,
"ColorG": 173,
"ColorR": 57,
"IntervalsFrom": [
8,
89,
170
],
"IntervalsTo": [
25,
106,
187
],
"Name": "Images in RVF",
"NrOfIntervals": 3
},
{
"ColorB": 146,
"ColorG": 91,
"ColorR": 59,
"IntervalsFrom": [
62,
143,
224
],
"IntervalsTo": [
79,
160,
241
],
"Name": "Images in BVF",
"NrOfIntervals": 3
}
],
"FileName": "CG_OBJECTS.prt",
"Name": "Untitled",
"NrOfConditions": 4,
"TimeCourseColors": {
"RefTCPenThick": 3,
"RefTimeCourseB": 0,
"RefTimeCourseG": 255,
"RefTimeCourseR": 255,
"TCBackColorB": 0,
"TCBackColorG": 0,
"TCBackColorR": 0,
"TCPenThick": 3,
"TCTextColorB": 255,
"TCTextColorG": 255,
"TCTextColorR": 255,
"TimeCourseB": 255,
"TimeCourseG": 255,
"TimeCourseR": 255
},
"TimeResolution": "Volumes"
},
"SliceAcquisitionOrder": 3,
"SliceAcquisitionOrderVerified": true,
"SliceGap": 0.9900000095367432,
"SliceThickness": 3,
"SliceTimingTableSize": 0,
"TimeResolutionVerified": false,
"Version": 1,
"VoxelResolutionVerified": true
},
"ConversionSoftware": "BrainVoyager",
"ConversionSoftwareVersion": "21.0.0.3592",
"ImageNumber": 1,
"ImageOrientationPatientDICOM": [
0.99756405,
0,
-0.069756478,
0,
1,
-6.9388939e-18
],
"ImagePositionPatientDICOM": [
-561.97581,
-576.86747,
-15.446725
],
"ImageType": "ORIGINAL PRIMARY M ND MOSAIC",
"MagneticFieldStrength": 2.89362,
"Manufacturer": "SIEMENS",
"ManufacturersModelName": "Trio",
"Modality": "MR",
"NrOfTemporalPositions": 0,
"PatientPosition": "HFS",
"RepetitionTime": 2000,
"SeriesDescription": "ep2d_bold_tbv_std",
"SeriesNumber": 3,
"SiemensMosaicSliceAcqOrder": 0,
"SiemensMosaicTE": 0,
"SiemensMosaicTR": 0,
"SliceThickness": 3,
"StudyDescription": "RaiGoe^standaard"
}
TSV Sidecar File for protocol-related information
While the protocol information in the JSON sidecar of a functional (FMR) NIfTI file can be used directly by BrainVoyager, the BIDS standard requires that this information is stored in a special tab-separated value (TSV) text file containing the onset times and durations (in seconds) of all events/blocks of a functional run. In addition to the BrainVoyager-specific protocol information in the JSON file, BrainVoyager also saves protocol information (if available) in such a TSV file (see example below) so that other BIDS-compatible analysis software is able to use the protocol data. Note that the TSV file may also contain more columns for additional behavioral data such as information about button presses and reaction times.
Example TSV protocol sidecar file "sub-01_ses-01_task-images_run-01_events.tsv":
"Condition Name" Onset Duration
"Fixation" 0 14
"Images in RVF" 14 36
"Fixation" 50 18
"Images in LVF" 68 36
"Fixation" 104 18
"Images in BVF" 122 36
"Fixation" 158 18
"Images in RVF" 176 36
"Fixation" 212 18
"Images in LVF" 230 36
"Fixation" 266 18
"Images in BVF" 284 36
"Fixation" 320 18
"Images in RVF" 338 36
"Fixation" 374 18
"Images in LVF" 392 36
"Fixation" 428 18
"Images in BVF" 446 36
"Fixation" 482 18
The first row of a BIDS-compatible TSV file must contain the names of the information stored in the respective columns; each subsequent row contains the data for one event of the functional run. Note that in the example file above, tabs in the file are interpreted as tabs - in a more veridical representation one would see a single tab sign ('\t') between entries within a row.
Visualizing JSON and TSV files
As described above, BrainVoyager looks for JSON and TSV files when loading a NIfTI file to access useful information. If these sidecar files are found, they will be automatically displayed in appearing JSON and TSV viewer windows. It is also possible to view specific JSON and TSV files at any time by using the Open JSON and Open TSV items in the File menu. The screenshots below show the JSON and TSV files of the example functional data sidecars presented above. Since a JSON file is a hierarchical data structure (the value of a key inside an object can itself contain an object), the JSON data is presented in a tree view allowing to open and close the content of objects (visualized as sub-trees). In the example JSON file shown below, the "BrainVoyagerInfo" object has been opened, while the details of the "Protocol" object inside the "BrainVoyagerInfo" object are hidden (collapsed tree).
Copyright © 2023 Rainer Goebel. All rights reserved.