BrainVoyager v23.0
FA Using Boundary-Based Registration
Since BrainVoyager 20.2 boundary-based registration (BBR) is available as a powerful method to coregister anatomical data with other data sets from the same subject as long a they have sufficient grey/white matter contrast. One of the most important applications of BBR is the fine-tuning adjustment (FA) step when coregistering functional data of runs with an anatomical data set from the same scanning session. BBR can now be used instead of the other available approaches that are based on gradient or intensity-driven coregisttration. While BBR performs at least as good as these other alignment approaches, some preparatory steps are required. These preparatory steps include white/grey matter segmentation of a VMR data set, reconstruction of a WM-GM cortex mesh from the segmented brain and eventually a spatial transformation step (for more details, see topic Boundary-Based Registration). In the following sections the necessary steps in the context of FMR-VMR coregistration are described. In case functional data is analyzed in FMR-VTC space (e.g. when processing high-resolution data), the preparatory steps are different and described in topic VMR to FMR-VTC Space Alignment Using BBR. The steps for standard FMR-VMR alignment using BBR are:
- Creation of a whole-cortex mesh in native space from an anatomical data set recorded in the same session as the functional (or diffusion-weighted) data. This involves the standard intensity-inhomogeneity step (including skull stripping) and an (automatic) segmentation step.
- Creation of a functional volume stored in a VMR file in a space close to the native space anatomical VMR. This is achieved by running a normal FMR-VMR alignment procedure except that only the initial alignment (IA) step is necessary. Since this step uses header information from the functional and anatomica data, the created functional volume (see berlow) will be close enough to the native space VMR allowing the BBR process to perform the fine-tuning adjustment step. The IA step will also create an IA transformation matrix (_IA.trf file) that will be one piece of input for the creation of VTC data.
- The created functional volume (VMR) created in step 2 serves as the host VMR of the whole-cortex mesh created in step 1. The BBR process is started from the surface (OpenGL) window containing the cortex mesh aligning the mesh to the functonial volume. As a result, an FA spatial transformation matrix (.trf file) will be created that can be used in the usual "Create VTC" step.
- The functional data (with performed preprocessing) is transformed in native, ACPC, TAL or MNI space in the usual way using the "Create VTC" dialog. The initial alignment (IA) matrix file created in step 2 is used for the IA step (as usual) and the fine-tuning (FA) alignment matrix from step 3 is used to optimally align the functional and anatomical data.
In the following sections, these steps are described in more detail. Note that steps 1-3 are performed automatically when chosing BBR in the FMR-VMR coregistration sub-workflow in the data analysis manager.
Native-Space Cortex Mesh from Intra-Session Anatomical VMR
BBR uses a cortex mesh (SRF) as input which is used to align the respective anatomical data (VMR) with another data set. To obtain a suitable cortex mesh, the underlying anatomical data set (VMR) needs to be segmented. Since the FMR data is aligned usually ot the intra-session anatomical data set, this VMR file can be used for the segmentation process. It is also possible to first transform the intra-session VMR into ACPC (or TAL) space, perform standard (advanced) segmentation and afterwards transforming the segmented brain back into native space. In the following description, the direct segmentation in native space is described:
- Before the segmentation can be started, the intra-session VMR needs to be created; note that the anatomical data set need to have good white/grey matter contrast, otherwise the cortex segmentation will produce suboptimal results.
- The intensity inhonogeneity correction (IIHC) step should be run next in order to remove the skull and to enhance the homogeneity of white (and grey) matter.
- The segmentation can be performed using the Automatic Cortex Segmentation For BBR dialog that can be invoked by clicking the Automatic Cortex Segmentation For BBR option in the Volumes menu. The new Automatic Segmentation for BBR dialog uses routines from the advanced segmentation pipeline without assuming normalized space, i.e. it can be run directly from a native space VMR; for more information, consult the topic Automatic Cortex Segmentation For BBR.
Functional Volume in Near-Native Space: Header-Based IA
In the second preparatory step, a volume (VMR) representation of the respective run of the functional (or diffusion-weighted) data needs to be created in a space that is close to the native space VMR. This is achieved by running the usual header-based initial alignment (IA) routine (without the FA step). Besides creating the IA transformation matrix, the header-based alignment step creates and saves a functional volume that is in a space close enough to the native space VMR so that the BBR routine is able to find a fine-tuning adjustment (FA) transformation that optimally aligns the two two data sets. In more detail the following steps are performed:
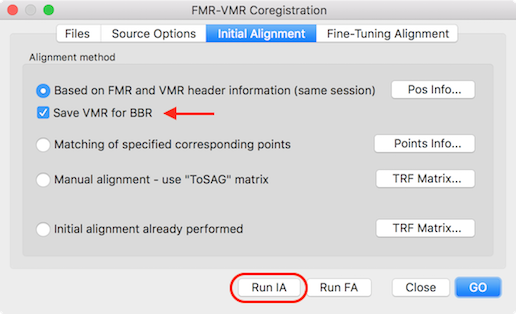
- Run the standard header-based initial alignment (IA) from the FMR-VMR Coregistration dialog that is invoked, as usual, from the Coregistration tab of the 3D Volume Tools dialog.
- As usual, select a functional data set from the same scanning session as the native-space VMR. Make sure that the selected FMR has not been spatially smoothed, i.e. it is recommended to select the original "first-vol" FMR file.
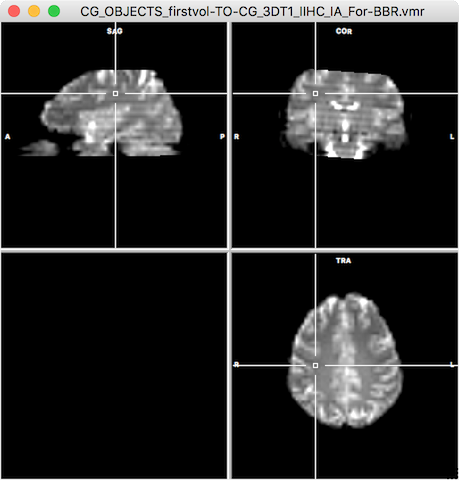
- Make sure that the new Save VMR for BBR option is checked in the Initial Alignment tab of the FMR-VMR Coregistration dialog (on as default, see snapshot above). After the header-based alignment step has been completed, a VMR file will be created and saved to disk in the same folder where the functional data set has been selected for the IA step (see snapshot below). The name of the created functional volume contains the name of the selected functional file (without the .fmr extension) followed by the substring "-TO-" followed by the name of the target native space VMR file (without the .vmr extention) followed by the substring "IA_For-BBR.vmr" (see title bar in snapshot below).
Running BBR in Native Space to Obtain FA Matrix
- Load the created volume of the functional data that will be in nearly the same space as the anatomical (mesh) data set.
- Open the created cortex mesh in native space from the functional volume (see snapshot of functional volume for example data above).
- Open the Boundary-Based Registration dialog by clicking the Boundary-Based Registration item in the Meshes menu. Click the GO button to start BBR, which will produce the spatial transformation FA TRF matrix file that can be used for VTC creation (see below).
The registration can be usually directly started using the default settings in the dialog. For more details about the dialog, see topic Boundary-Based Registration.
Creating VTC Data Using BBR FA Matrix
With the created FA spatial transformation matrix, the functional data can now be created as usual using the Create VTC dialog. For the IA transformation the IA .trf file need to be selected that has been created in the header-based alignment step described above. For the FA spatial transformation matrix, the .trf file created using BBR is provided as input. With these files, the full functional data can be created in native space (see below).
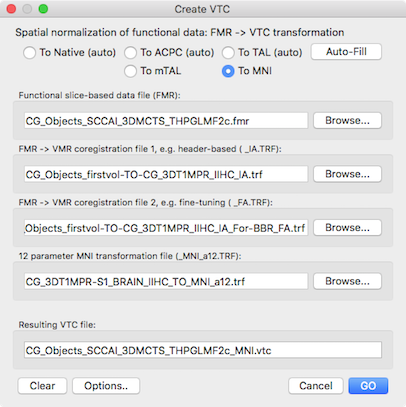
As usual, the functional data can also be further transformed in one step into normalized space (ACPC, TAL, MNI) in case that additional transformation files are available and provided in the Create VTC dialog. In the snapshot above, the functional data is further transformed into MNI space.
Multi-Run Coregistration
When using BBR for FMR-VMR alignment, it is recommended to align each individual run of a scanning session to the native VMR data set and to use the produced FA matrix for the respective run in the Create VTC dialog. It is thus not necessary (and not recommended for BBR) to use across-run motion correction since the FA transformation files produced by BBR for each run will handle the run-to-run displacements caused by head motion.
Copyright © 2023 Rainer Goebel. All rights reserved.