BrainVoyager QX v2.8
Cross-VTC Intersubject Correlation
In a standard GLM analysis, a single design matrix is created, which is fitted to the time course of each voxel. In order to test how similar the brains of subjects respond to the same stimulation, the time courses of a voxel of one subject can be modeled by the time course of the homologue voxel of another subject resulting in a voxel-by-voxel correlation map. When using the GLM approach, the time course of a voxel of a "source" subject is used to create one predictor of a design matrix to model the time course of the same voxel of the "target" subject. Thus, each voxel gets its own design matrix consisting of the time course of the "source" subject. In addition to the source voxel's time course, a constant is added as an additional confound predictor to complete the design matrix with two predictors. When the source time course and the target time course is z-normalized, the resulting beta value for the source time course predictor can be directly interpreted as the correlation value between the voxel's source and target time course.
In order to be meaningful, the different subjects have to experience the same stimulation, e.g. by using the same stimulation protocol. Note, however, that the protocol need not be explicitly specified. Instead of a classical stimulation protocol, the innovative fMRI study by Hasson et al. (2004) scanned subjects viewing the same movie during individual scanning sessions. When correlating the time courses from homologue (Talairach) voxels from different subjects, they observed highly synchronized responses in widespread cortical networks. The example below uses a traditional localizer experiment with faces, houses and object stimuli presented in the same order to all subjects.
Application
To perform intersubject correlation, load the Talairach-normalized VMR data set of the target subject and link the time course (VTC) data of that subject. This time course data provides the to-be-modeled data for each voxel. Enter the Single Study General Linear Model dialog and click Options to invoke the Single Study GLM Options dialog. Finally click the Residuals button in the lower left corner to invoke the Serial Correlations Options dialog (see snapshot below).
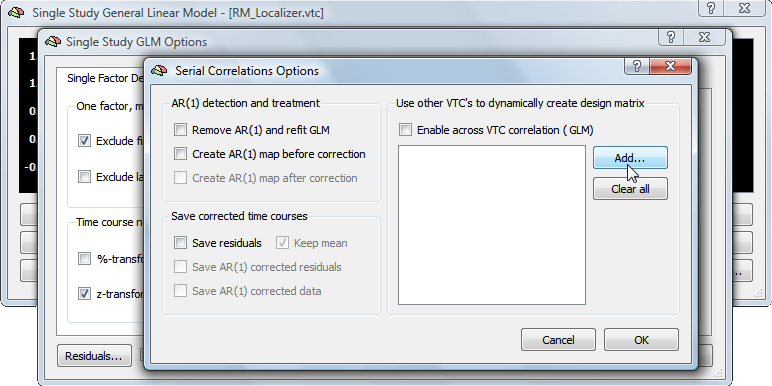
Click the Add button in the Use other VTC's to dynamically create design matrix field and browse to the VTC data of another subject. Make sure that the Enable across VTC corelation (GLM) option is checked, which should have been turned on automatically when selecting the source VTC data. The snapshot below shows the state of the dialog after the selection of a source VTC file.
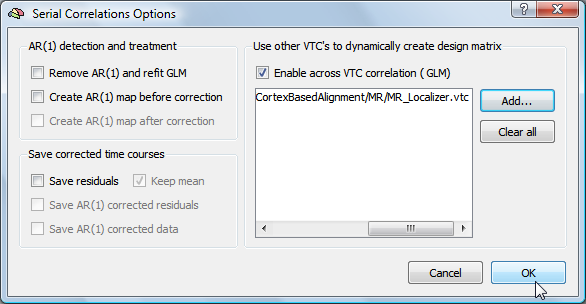
Note that it is possible to select additional VTC files from other subjects; the time course of each VTC will then be added to a voxel's design matrix. Note, however, that this option should only be used when the goal is to detect unique contributions from other subjects. For the purpose of investigating the similarity between two subjects, the procedure described above should be repeated for each "source" subject.
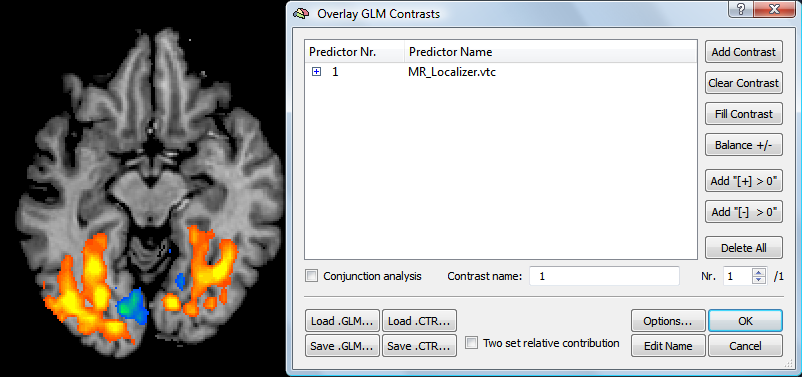
After specification of the source VTC file, close the Serial Correlations Options dialog by clicking the OK button. Also close the Single Study GLM Options dialog by pressing its OK button. Then click the GO button in the Single Study General Linear Model dialog. A progress bar will appear providing feedback about the creation of the voxel-by-voxel cross-VTC correlation map. The snapshot below shows the result for a Localizer experiment showing strong intersubject modulation.
References
Hasson et al (2004). Intersubject synchronization of cortical activity during natural vision. Science, 303, 1634-1640.
Copyright © 2014 Rainer Goebel. All rights reserved.