BrainVoyager QX v2.8
FA Using Intensity-Driven Rigid Transformations
In the case of a simple sagittal reorientation of the functional data, the functional and anatomical volumes might still be displaced substantially and the automatic detection of the correct translation and rotation parameters might be difficult. To be able to find the right parameters also in the case of large displacements, this alternative fine-tunibg alignment approach uses a coarse-to-fine alignment procedure, computing a Gaussian pyramid for each volume containing several down sampled (low-resolution) versions of the original data sets. The intensity-driven alignment procedure then works first on the low-resolution representations to correct large displacements without being trapped in local minima. After the alignment at a coarse scale, the alignment continues at progressively finer resolutions. The end-result is a high-quality alignment of the functional and anatomical data. Current experience shows that this procedure works fine if the VMR and FMR data set does not contain intensity inhomogeneities across space. In case of inhomogeneities across space, the fine-tuning adjustment might push the data set a few millimeters off the optimal result. We advice, thus, to run inhomogeneity correction of the VMR data set prior to using this function in case that the observed alignment is not very accurate. FMR data sets normally do not suffer strongly from inhomogeneities.
Note. The descriptions below need only be consulted in case of problems with the recommended gradient-based fine-tuning alignment since it produces better results in most cases.
After the initial alignment step has been performed, you can start the intensity-driven multi-scale alignment step. Invoke the FMR-VMR Coregistration dialog by clicking the Align button in the FMR-VMR coregistration field of the Coregistration tab of the 3D Volume Tools dialog. Switch to the Fine-Tuning Alignment tab. Since the intensity-based multi-scale alignment is not the default mode for the fine-tuning coregistration step, the Intensity alignment using multi-scale approach option must be clicked to select it (see snapshot below).
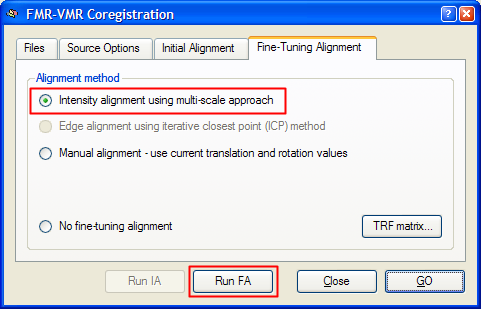
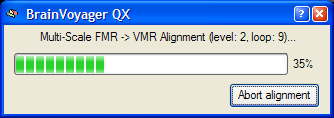
To start the alignment, click the Run FA (or GO) button. During the alignment procedure, a dialog informs about the progress made (see snapshot below). The alignment procedure lasts about a minute and includes a preparation step for the creation of the multi-resolution pyramids, and multiple parameter adjustment loops for resolution levels 2 and 1. If the dialog reaches 99% but does not close automatically, you can press and hold the Escape button to stop the alignment process.
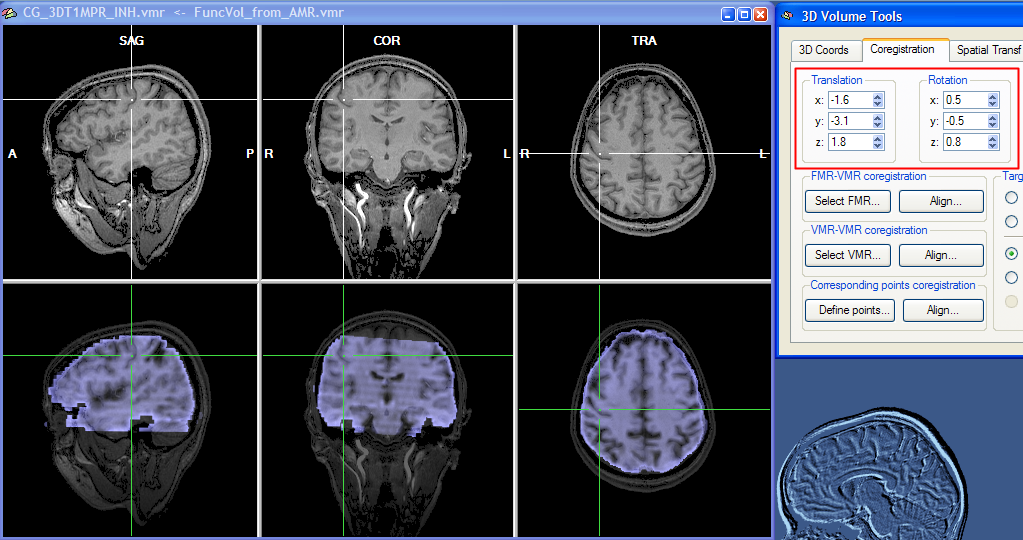
You can observe the iteratively updated parameter estimates during alignment in the Coregistration tab of the 3D Volume Tools dialog. The snapshot below (red rectangle) shows the estimated parameters in the Translation and Rotation field after the fine-tuning adjustment has been completed. The lower row of the VMR window shows the functional and anatomical volume in transparency blend mode. Details of different visualization options for the source and target volume can be found in the IA using header-based coregistration topic.
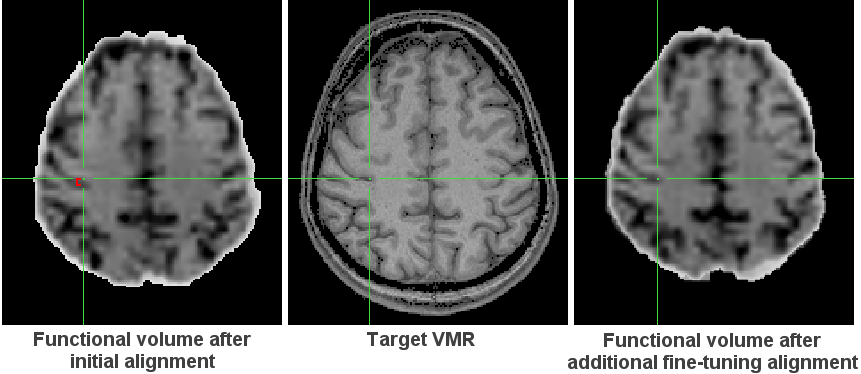
The estimated parameters show that in our example data, the most substantial improvement over the initial alignment step has been achieved by a translation of -3.1 mm along the Y axis, i.e. along the anterior - posterior direction. The following images highlight this offset (see red marker and position of green cross) comparing the result after the initial alignment (left image) with the result of the additional fine-tuning alignment (right image) with respect to the target 3D anatomical volume (middle image). Such comparisons also for other brain regions confirm the improvement achieved by the fine-tuning alignment step.
Copyright © 2014 Rainer Goebel. All rights reserved.