BrainVoyager QX v2.8
From Measured Slices to 3D Space
While basic analysis of DWI data can be performed in measurement (DMR-DWI) space, advanced applications, such as group analysis of FA maps and fiber tracking, require that the data is transformed into (normalized) 3D space. This includes a coregistration step aligning the DMR-DWI data to a 3D VMR data set and, optionally, a transformation step into a standard 3D space, e.g. AC-PC or Talairach space.
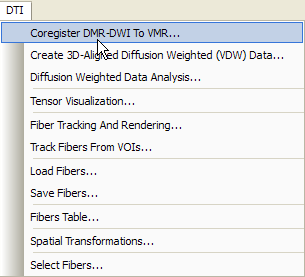
The steps to align the DWI data with a high-resolution 3D structural scan of the subject are identical to those described for coregistration of FMR and VMR data sets. After loading a 3D data set (VMR file) measured in the same session as the DWI scan, the alignment routines can be invoked by clicking the Coregister DMR-DWI To VMR item in the DTI menu (see snapshot above).
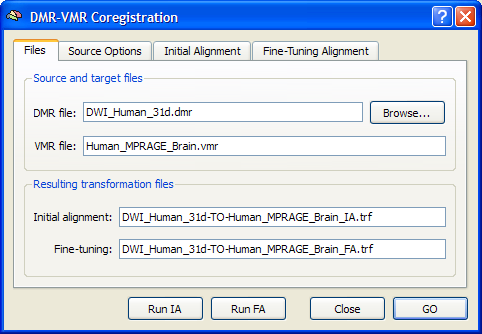
In the appearing DMR-VMR Coregistration dialog, click the Browse button and select the DMR file to be coregistered to the loaded VMR file. To run the header-based initial alignment, click the Run IA button. In the VMR view window check by visual inspection that a good initial alignment of the DMR-DWI data and the VMR data has been obtained (for details, consult the header-based initial alignment documentation). Then reenter the DMR-VMR Coregistration dialog and click the Run FA button to further improve the coregistration result. You may inspect the estimated translation and rotation parameters of the fine-tuning alignment step in the Coregistration tab of the 3D Volume Tools dialog.
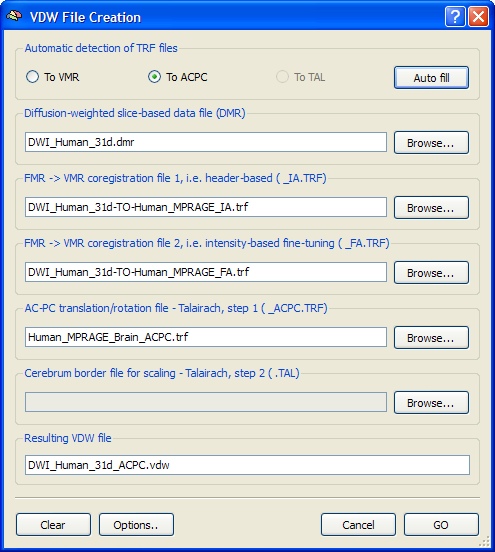
After coregistration of the DMR-DWI data with the intra-session 3D VMR data set, it can be transformed into the space of the VMR data. This transformation produces a VDW (volumes of diffusion weighted data) data set, which is saved to disk. A VDW file is similar to a VTC file except that it contains appropriate header information about the gradient table and a history of applied spatial transformations in order to allow correct interpretation of diffusion directions in the new 3D space. While the intra-session 3D space allows to calculate additional diffusion maps and fiber tracking, group analyses of FA maps requires transformation into Talairach space. In order to transform the DMR-DWI data to ACPC or Talairach space, the respective ACPC transformation file and eventually Talairach landmark file must be created for the intra-session VMR data set as described in the Brain Normalization topics. If all files for the DMR-DWI to VDW transformation are available, enter the VDW File Creation dialog by clicking the Create 3D-Aligned Diffusion Weighted (VDW) Data item in the DTI menu. The snapshot above shows an example for creation of VDW data in ACPC space; after selecting the source DMR project file, the To ACPC option has been turned on and the remaining entries were automatically filled-in by clicking the Auto-Fill button.
As default, the voxel resolution of the created VDW data is 2 mm. If you want to create the data in higher or lower resolution, enter the Create VDW Options dialog by clicking the Options button and select the 1x1x1 or 3x3x3 option. As default, the data is resampled using sinc interpolation in order to produce 3D space data with highest quality. Note that sinc interpolation is slow and it may take several 10s of minutes to transform a single DWI data set with the Sinc option. While not recommended for DWI data, you may switch to the Trilinear option in the Interpolation options field; this option also produces good results but introduces slight spatial smoothing.
When the program has finished creating the VDW data, diffusion tensors can be estimated in the respective 3D space.
Copyright © 2014 Rainer Goebel. All rights reserved.