BrainVoyager QX v2.8
Reconstruction of Cortex Hemispheres
While the output from the GM-CSF tab with labelled grey and white matter is sufficient for subsequent cortical thickness measurements, the obtained segmentations can also be used to create high-quality cortex reconstructions to visualize high-resolution functional data that has been aligned with high-resolution anatomical data with high precision on high-resolution cortical surface meshes. While this can be obtained with standard segmentation and reconstruction tools, the Hemispheres tab of the Advanced Segmentation Tools dialog (added in BrainVoyager QX 2.8) helps to separate cortical hemispheres and to produce optimally smoothed reconstructed surface meshes.
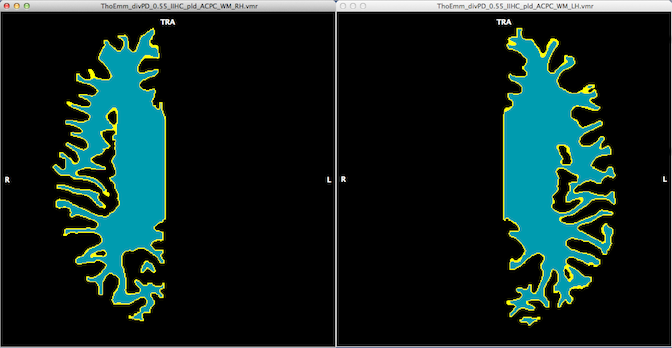
The program assumes that a "_WM_GM.vmr" file resulting from the previous steps is available, i.e. just obtained by running the tools in the other tabs or loaded from disk. In order to disconnect the whole-cortex WM_GM data set, the Disconnect button in the Disconnect hemispheres field needs to be clicked. Before clicking the button, the progam already presents the two file names under which the two segmented hemispheres will be stored. Note that this function not only disconnects the two hemispheres but it also preapres the white matter segments for cortex reconstruction as can be seen by the resulting two data sets that have been loaded from disk and shown in the snapshot below.
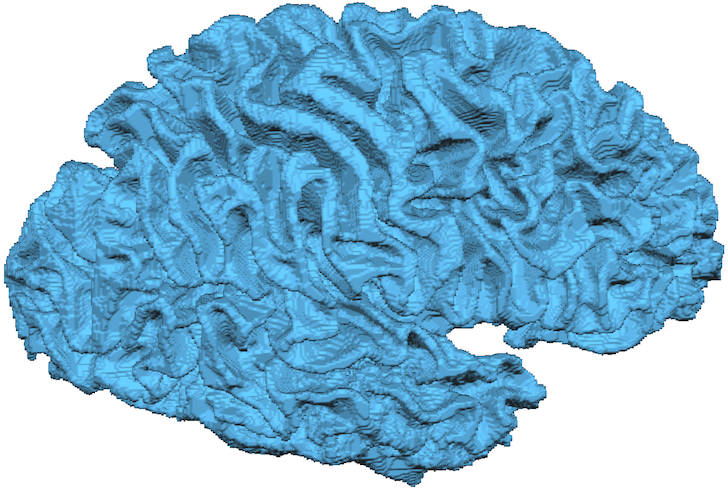
To create reconstructed cortex meshes from each prepared white matter hemisphere file, the Reconstruct button in the Reconstruct cortex meshes from prepared hemispheres field needs to be clicked next. The program then uses the previously stored prepared cortex mesh files and produces two mesh files each containing the reconstructed (unsmoothed) border of the white matter voxels. The names of the mesh files that will be stored when running this function are shown in the Mesh LH file and Mesh RH file text boxes. For the snapshot below, the resulting right hemisphere "RECO" mesh has been loaded from disk. Note that the mesh reconstructed from 0.55 mm voxels looks rather smooth as compared to reconstructed meshes from standard 1 mm segmentations (see also below).
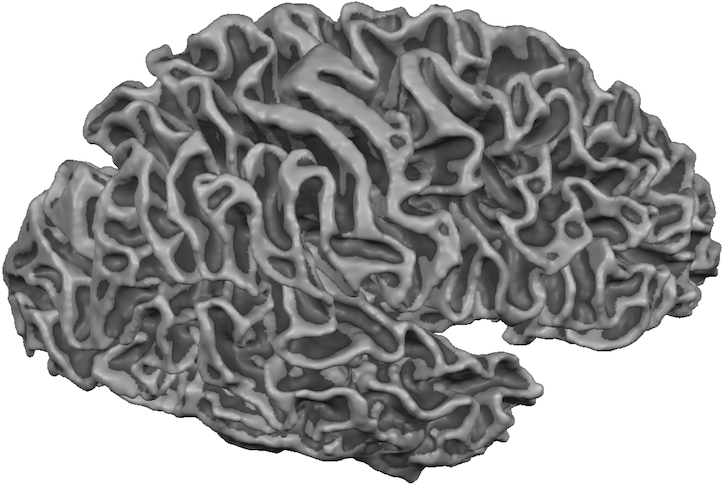
The program will also create smoothed versions of the reconstructed meshes in case that the And Smooth option is turned on (default) in the Reconstruct cortex meshes from prepared hemispheres field. The resulting "RECOSM" mesh for the right hemisphere has been loaded from disk for the snapshot below. Note that the program also creates curvature colors automatically besides smoothing the mesh. Further note that the program uses advanced mesh smoothing during this process that has been introduced in BrainVoyager QX 2.8. This advanced "reco" smoothing only smoothes high-frequency curvature such as the jags resulting from 90 degree voxel border edges but it does not produce more global "shrinking" shape effects as introduced with conventional smoothing.
The last function in the Hemispheres tab allows to create versons of the "WM_GM" file and especially prepared left and right matter segmentations in standard 1 mm (256 dimension) space by clicking the Downsample button. This can be useful in case that one wants to use advanced segmentation tools but continue with further analysis with less "big" data files, e.g. for visualization and analysis of fMRI data of conventional resolution (2-3 mm). Furthermore, the bridge removal tool currently only operates for 1 mm space data sets. Note that advanced segmentation, including the downsampling step, are now also directly integrated in the standard automatic segmentation pipeline as an option. The file names of the resulting downsampled files are shown in the Downsampling to 1.0 mm resolution field. The resulting files are distinguished from their high-resolution counterparts by the addition of the "_DS1mm_" substring in the stored file names.
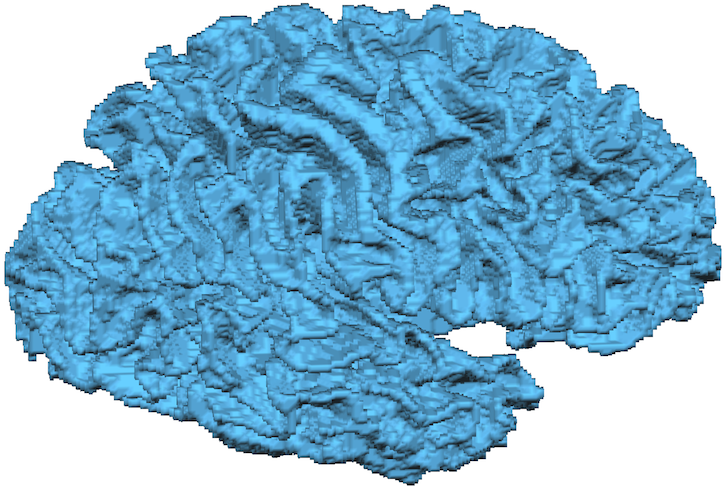
The downsampling function does not automatically reonstruct the meshes. In order to get the right hemisphere cortex mesh shown in the snapshot above, the prepared "_WM_RH" white matter segmentation file was loaded and then reconstructed using the Create Mesh dialog. While the mesh looks similar as the high-resolution version (see aboe), it is evident that the larger voxels lead to a less-smooth visual appearance of the "RECO" mesh. After smoothing the mesh (click on the Morph Mesh icon), the resulting mesh from 1 mm voxels looks very similar as the one created rom 0.55 mm voxels (see snapshot below and compare to corresponding high-res snapshot above).
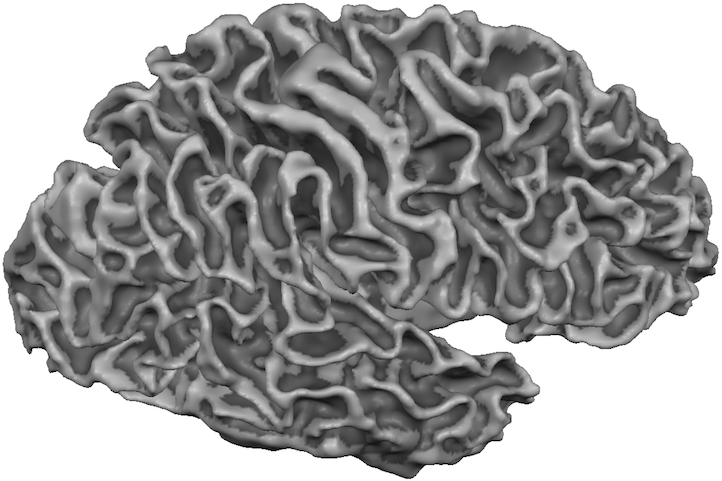
Note that the downsampled white matter segmentations are not the result of a volume downsampling process (only the "_WM_GM" file is created by volume downsampling) but are created by projecting the high-resolution surface mesh into the 256 (1 mm) space. While this would only produce voxels at the white/grey matter boundary, the interior is filled subsequently to create a white matter segmentation. This approach leads to more preceise surface meshes than using a volume downsampling.
Copyright © 2014 Rainer Goebel. All rights reserved.