BrainVoyager QX v2.8
Single-Factor Repeated Measures ANOVA
The ANOVA model with one within-subjects factor is an important basic model suitable for many single-group fMRI designs. It assumes that each subject has received all experimental conditions (repeated measures). In the following, it is first described how to use the ANCOVA dialog to run this model over all voxels (or vertices) in order to obtain statistical random effects (RFX) maps and how the factor main effect as well as specific contrasts can be tested. Then it is described how the ANCOVA dialog can be used to apply the one-way within-subjects factor model to any region-of-interest (ROI). The documentation in this section uses a simulated data set, which can be generated by running the "Group Data Simulator" plugin with the "Design_1W.gds" file (for details, see the online help of the plugin).
Providing a RFX-GLM as Input
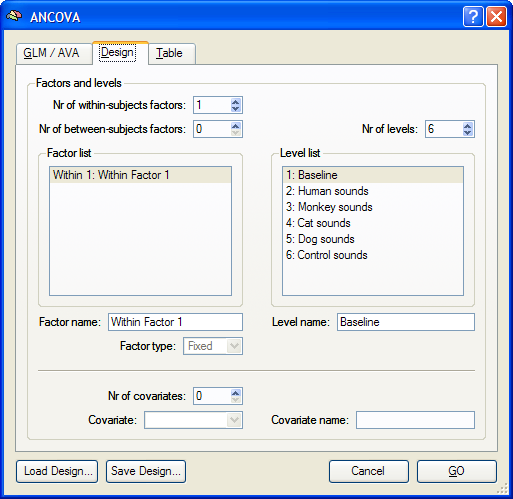
In order to run an ANOVA analysis for this design, open the ANCOVA dialog from the Analysis menu. If you continue right away from a computed RFX-GLM, the beta values are already available and the design can be specified in the displayed Design tab. If no appropriate GLM data structure is available, select a previously calculated RFX-GLM file in the GLM / AVA tab. The program then automatically switches to the Design tab representing initially always a design with one within-subjects factor, which is listed in the Factor list. The levels of this "Within 1" factor are shown in the Level list containing all conditions (names of beta values) found in the provided GLM.
Baseline Condition?
In versions prior to BrainVoyager QX 2.4, a baseline condition was included when starting from a GLM data file. While usually ignored when defining the design, the baseline was recommended for models with only one main condition (e.g. "Stim" vs "Rest"). Since the baseline condition is not optimal for running ANOVA models (no independent variance estimation), the baseline beta is now dropped when opening a GLM file in the "ANCOVA" dialog. For designs with only one main condition (and one group), the 1-sample t-test model is appropriate and will be automatically suggested.
Specification of the Design
Since this design is the default model of the ANCOVA dialog, the number of within-subjects (1) and between-subjects (0) factors need not be changed. The number of levels and corresponding condition names are also determined automatically from the provided RFX-GLM and have been put in the Level list. The number of levels for this design can not be changed at present, but the name of each level can be changed, if desired. To change the name of a level, select a level in the Level list and then change its name in the Level name text box. Although not necessary, it is also useful to edit the name of the factor in the Factor name field (e.g. from "Wihtin Factor 1" to "Sounds" for the sample data used in this description). To run the single-factor ANOVA, click the GO button. The resulting ANOVA results for each voxel are stored in the AVA file specified in the GLM / AVA tab.
Testing Effects and Contrasts
The single-factor repeated measures ANOVA model allows testing an overall main effect (F test) as well as specific contrasts comparing mean condition values (t tests), e.g. whether the effects (beta values) of two conditions differ significantly from each other. After calculating the ANOVA model, the overall F map is shown as default. Other tests can be selected in the Overlay RFX ANCOVA Tests dialog (see snapshot below), which can be invoked by selecting menu entry Overlay RFX ANCOVA in the Analysis menu. If this dialog is called and no AVA file is available, most options are disabled. In that case, the Load AVA button can be used to select the AVA file. The program tries also to load the original RFX-GLM file from which the AVA file has been calculated. While this GLM file is not necessary for overlaying ANOVA tests, it allows to show the Voxel Beta Plot dialog with the data (beta values) at the voxel under the mouse cursor.
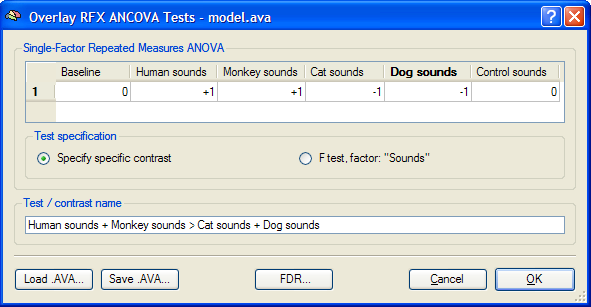
The Overlay RFX ANCOVA Tests dialog (see snapshot above) contains a contrast table showing the levels of factor A (within-subjects factor "Sounds" in the example design) in the top row. The entries in the cells of the table can be changed to test specific contrasts comparing specific levels (cell means). The options in the Test specification field allow to select the type of test, which can be a specific contrast (Specify contrast option, resulting in a t map) or the F test for the within-subjects factor (F test, factor: <name> option). As default, multiple comparisons correction will be turned off for the resulting statistical (F or t) map but can be turned on using the FDR button. Clicking the OK button will calculate and overlay the selected test as a statistical map on the underlying anatomical data set (VMR or SRF). In the snapshot above, a contrast test has been selected. The entries in the cells of the contrast table can be changed from "0" to "+1", "-1" and "0" by clicking repeatedly on a chosen cell. Double-clicking a cell allows entering a numerical value directly. The specified contrast in the snapshot above tests the contrast "[Human sounds + Moneky sounds] > [Cat sounds + Dog sounds]". For the example data, the Voxel Beta Plot is shown below for two voxels, one from a voxel of "VOI 1" and one from a voxel of "VOI 2". The response pattern in a voxel of "VOI 1" shows that this region responds much stronger to human (red disks) and monkey (green disks) sounds than to cat (dark blue disks) and dog (light blue disks) sounds, while the reverse pattern is evident for "VOI 2" (see snapshots below).
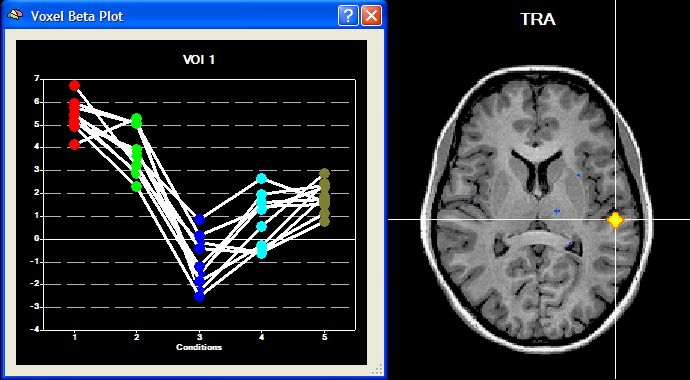
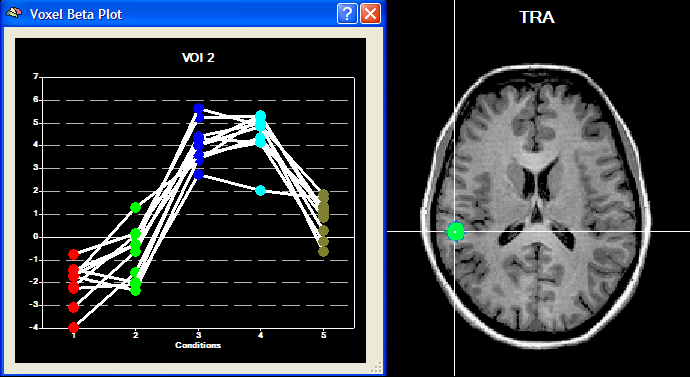
Testing the Model for ROI Time Courses
Besides calculating statistical ANOVA maps (VMPs or SMPs), it is possible to run the ANOVA analysis for any region-of-interest (ROI) providing detailled numerical output. As a prerequisite, VOIs have to be available, which can be loaded from disk or directly defined from functional or anatomical data. After functionally defining the two VOIs "VOI 1" and "VOI 2" for the sample data, the ANCOVA dialog can be invoked from the VOI Analysis Options dialog, which itself can be called from the Volume-Of-Interest Analysis dialog. Before calling the ANCOVA dialog, the same multi-subject design matrix file (e.g. "Sounds.mdm") used to calculate the RFX-GLM has to be selected in the Design matrix file text box of the GLM Options tab. When running the ANOVA, the MDM design matrix file is used to run a first-level RFX-GLM over the VOI time courses of all subjects in order to obtain VOI beta values as input for the second-level ANOVA analysis. After the MDM file has been specified, a VOI can be selected in the VOI list of the VOI GLM tab. The ANCOVA dialog can be launched from the VOI GLM tab by clicking the ANCOVA button in the VOI RFX analysis field. The ANCOVA dialog will show two tabs, the Design and the Table tab. Since the default design corresponds already to the desired one within-subjects factor model, nothing has to be changed. It might be, however, helpful to change the factor name (and eventually level names) as described above. The table in the Table tab contains the estimated VOI beta values estimated by internally running a separate-subjects (RFX-)GLM over the VOI time course data of each subject.
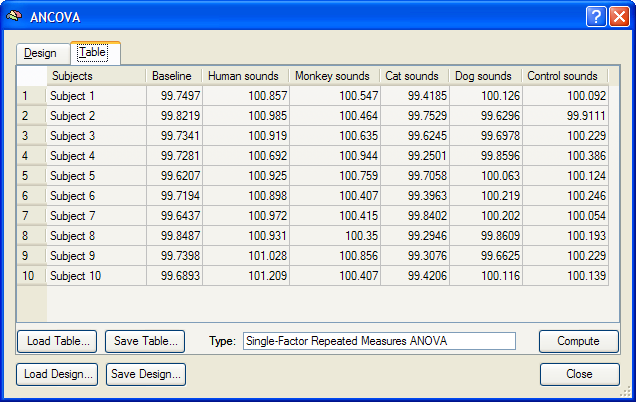
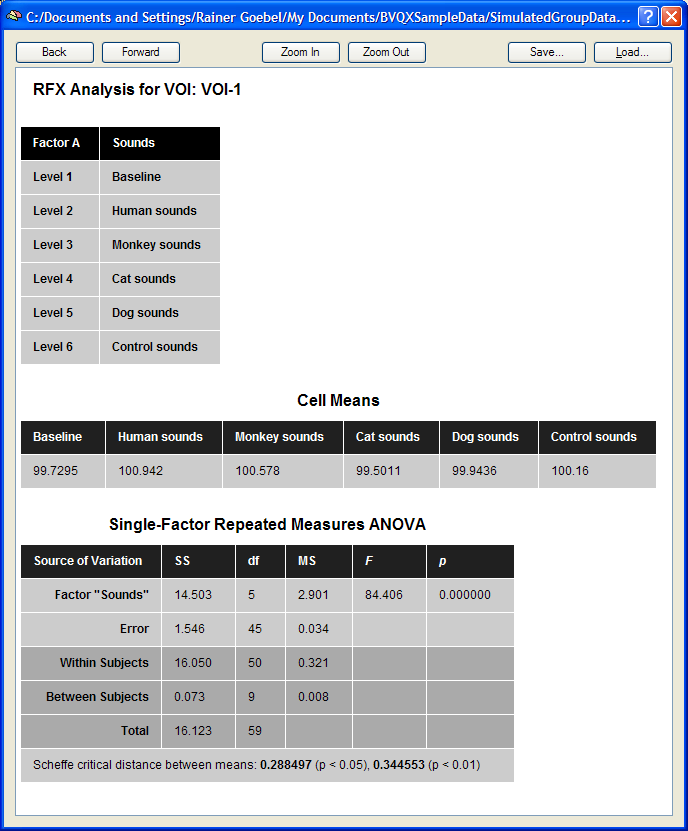
The single-factor ANOVA model can be calculated by clicking the Compute button in the Table tab. The results are presented in tabular form in three sections as a HTML document (see snapshot below). The first section defines information about the factor and its levels. The second section, "Cell Means", shows the mean values for all factor levels (mean condition effects in ROI). The third section shows the ANOVA table. The most important information of this table is the F test indicating whether there is a significant overall effect. The Scheffe critical difference indicates how much cell values must differ from each other in order to be significantly different.
Validation of Calculations
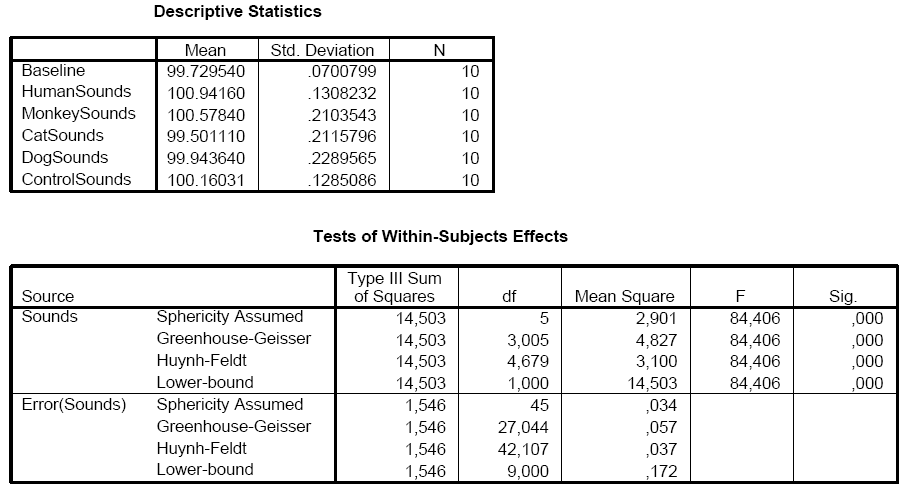
The VOI data of the table in the Table tab of the ANCOVA dialog can be saved to disk using the Save Table button allowing further analysis, if desired. During development of the ANCOVA module, the saved table data has been analyzed in SPSS 15 to test that the implemented calculations in BrainVoyager QX are performed correctly. As the snapshot below shows, the results from the SPSS analysis are identical to the ones obtained in BrainVoyager QX. The "Descriptive Statistics" shows that the calculated mean values correspond to the values in section 2 ("Cell Means") of the BrainVoyager QX output. The SPSS results in section "Tests of Within-Subjects Effects" are identical to the results obtained for factor "Sounds" and the associated error.
Copyright © 2014 Rainer Goebel. All rights reserved.