BrainVoyager QX v2.8
Independent Component Analysis (ICA)
Independent Component Analysis (ICA) is a data-driven method to analyze fMRI data. For fMRI data, spatial ICA (sICA) is normally applied as opposed to temporal ICA. Without further reduction of the data dimensionality, spatial ICA produces as many components as there are data points in the processed time course (VTC) data. There exist different ICA algorithms to estimate a set of spatial components. In BrainVoyager QX, the spatial decomposition of the data is performed using "FastICA", a fixed-point ICA algorithm developed by Hyvarinen and colleagues (for details, see http://www.cis.hut.fi/projects/ica/fastica/). The FastICA algorithm minimizes the mutual information of the components using a robust approximation of the negentropy as a contrast function and a fast, iterative (nonadaptive) algorithm for its maximization. There are two ways to run FastICA, the deflation approach (default, recommended) and the symmetric approach. Before applying spatial ICA, the temporal dimension of the data set may be optionally reduced using Principal Component Analysis (PCA).
BrainVoyager QX allows to limit the ICA decomposition of a functional data set to those voxels, which are within a specified region with respect to the cortical sheet. For a multivariate technique like ICA, such a restriction to the relevant voxels not only reduces calculation time substantially but also typically improves the resulting decomposition by "focusing" the ICA to the relevant voxel time courses. The restriction is implemented by applying a cortical mask file to the VTC data, which can be derived easily after standard cortex segmentation routines. The term cortex-based ICA or cbICA (Formisano et al., 2004) is used to refer to spatial ICA restricted to cortical voxels.
The Independent Component Analysis Dialog
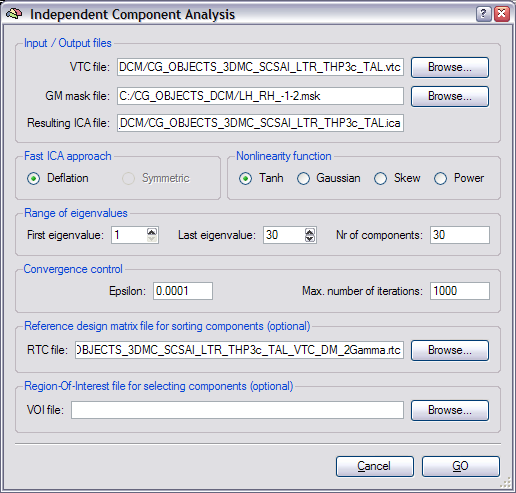
To run spatial ICA for a VTC data set, you may either click Independent Component Analysis in the Analysis menu or Independent Component Analysis (ICA) in the Plugins menu. In both cases, the Independent Component Analysis dialog appears allowing to specify details of the analysis. If a link to a VTC file has been already established for the current VMR prior to launching the dialog, the VTC file text box will be automatically populated with that time course file. You may, however, select any VTC file for analysis by clicking the Browse button on the right side of the VTC file text box.
The Overlay Independent Components Dialog
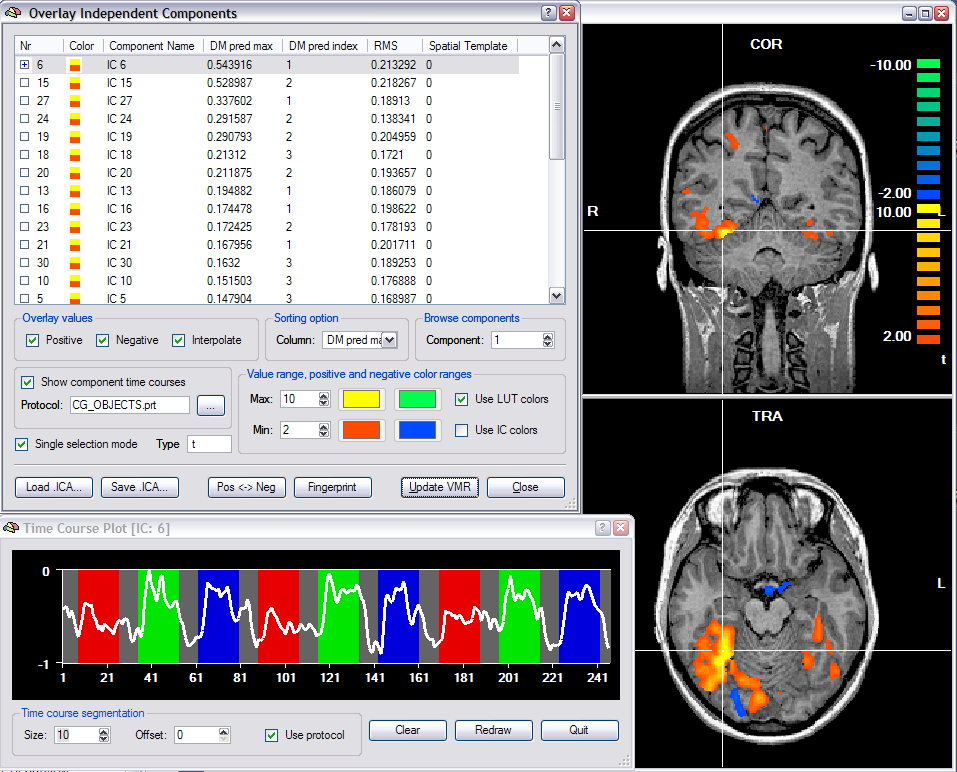
The actual ICA calculations are performed in a plugin, which has been implemented by Federico De Martino, Fabrizio Esposito and Rainer Goebel. This plugin is distributed with BrainVoyager QX and can be also downloaded from the BrainVoyager web site. The plugin saves a ".ica" file that can be loaded using the Overlay Independent Components dialog (see snapshot below) for visualization of component maps and associated time courses. Note that an ICA file is the same file format as a (native resolution) VMP file.
References
Formisano, E., Esposito, F., Di Salle, F. & Goebel, R. (2004). Cortex-based independent component analysis of fMRI time-series. Magnetic Resonance Imaging, 22, 1493-1504.
Copyright © 2014 Rainer Goebel. All rights reserved.