BrainVoyager QX v2.8
CBA Transformation of SMPs
When cortical hemispheres have been co-registered using cortex-based alignment, surface maps derived from a group-GLM are represented in group-aligned space since the group-GLM performs the CBA transformation implicitly for each subject's data using the provided transformation (SSM) files. Surface maps may be also created from other tools than with the GLM analysis tools, e.g. when running cortical thickness analysis (CTA). These tools run in "subject space", i.e. they are applied to (folded) SPH cortex meshes of individual subjects without application of SSM files. Several tools, including the ANCOVA tool and the creation of probabilistic maps expect surface maps as input that are already transformed into group-aligned space. The tools described in this topic allow to define associations between names of subject cortex meshes with corresponding SSM transformation files and to use the established associatons to apply the CBA transformation step to the included surface maps so that they will be in a common group-aligned space afterwards.
Preparing Group Surface Maps
As an example application of the presented tools, a probabilistic curvature map will be created before and after CBA transformation using curvature maps of cortex hemispheres that are created during the CBA procedure separately for each subject (i.e. they are in subject space).
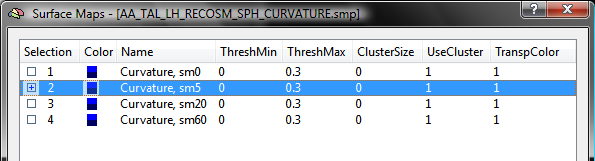
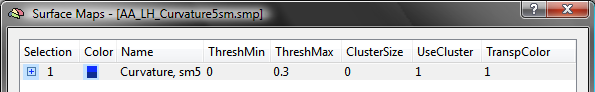
The snapshot above shows the curvature maps for the left cortex of the brain of one subject. For this example, the list of sub-maps is edited for each subject by removing all sub-maps except the one with smoothing level "5". The other 3 maps can be easily removed by highlighting them followed by pressing the Delete key. The resulting SMP has been saved to disk as the file "AA_LH_Curvature5sm.smp" (see below).
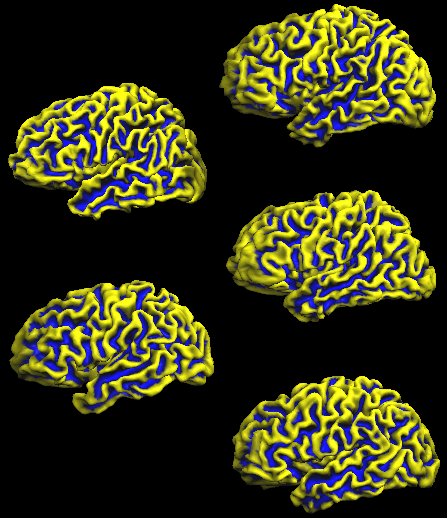
The same editing is then performed for the other four subjects used in this example resulting in 5 separate SMP files with curvature information defined in the space of the respective subject. The following figure shows the five individual folded SPH cortex meshes (left hemisphere) for all five subjects with the overlaid curvature map of the respective subject.
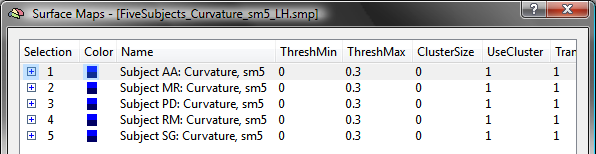
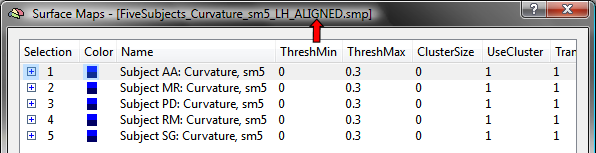
While individual maps could be CBA-transformed now, it is recommended to integrate the maps of each subject in a single "group" SMP file. This can be achieved by loading the SMP of the first subject using the Load SMP button in the Surface Maps dialog and then adding the maps of the other subjects using the Add SMP button. The resulting SMP data for the group of five subjects used in the example is shown in the snapshot below, which has been saved to disk as the file "FiveSubjects_Curvature_sm5_LH.smp". A necessary step for the application of CBA is that the maps of different subjects must be correctly identified; since the program uses the initial part of a map's name to identify a subject, the map's name must be edited to begin with the string "Subject [ID]: ". The "[ID]" part is replaced by the subject's identifier, which can be a number (e.g. "362"), text (e.g. "XY") or a combination of both (e.g. "S16"). In order to be consistent with the file names used during the CBA procedure, the same subject identifiers should be used also for the subject's surface maps. The name of a sub-map can be edited in the Surface Map Options dialog in the SMP Name field; the Surface Map Options dialog can be opened by using the Options button in the Surface Maps dialog.
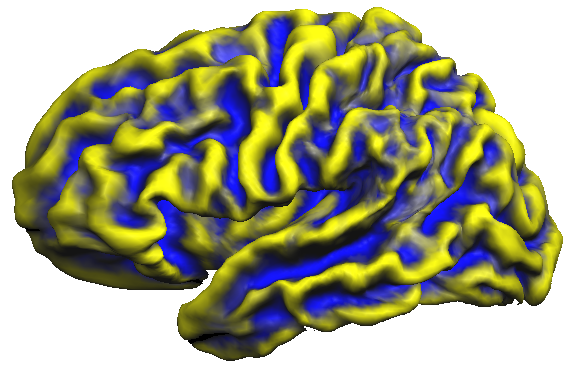
The snapshot below shows the accumulated curvature maps overlaid on the group-aligned folded cortex mesh. It is evident from the mixed yellow-blue color shades that concave regions indicating sulci (blue color) and convex regions indicating gyri (yellow color) are not overlapping very well. This is expected since the curvature maps are defined in subject space and are not transformed yet in group-aligned space.
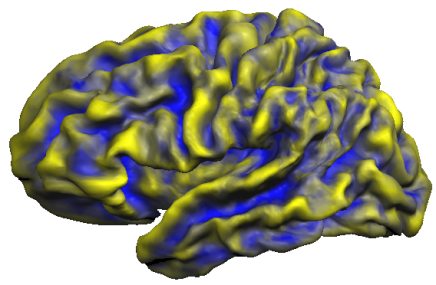
Another method to evaluate the consistency of maps across subjects is the calculation of probabilistic maps indicating how many subjects exhibit the same curvature information (convex or concave) at a given location (vertex).
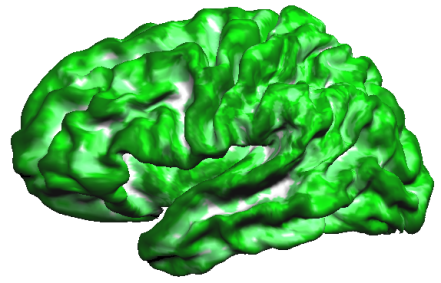
The resulting probabilistic map (see snapshot above) verifies that the alignment of the curvature maps in group-space is not very good in case that subject's curvature maps are not transformed using CBA information.
Linking Subject ID's with CBA Transformation Files
In order to transform the surface map of each subject with the correct CBA transformation file, the subject ID's must be associated with the matching SSM file. These associations can be performed using the Subject To CBA Data Assignment dialog, which can be invoked by pressing theCreate .S2S button in the Apply cb-alignment to maps created in subject space field of the Surface Map Options dialog (see snapshot below, yellow arrow). [Note that it is planned for a future release that ".S2S" files are automatically created when running the CBA procedure.] The procedure described here is identical to the one described in the CBA Transformation of POIs topic. If you have already created a .S2S file for POIs for the same group (and hemisphere), you can simply re-use that file by using the Browse S2S File button on the right side of the S2S File text field.
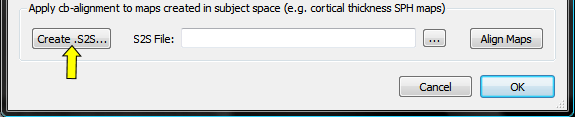
The Subject To CBA Data Assignment dialog (see snapshot below) contains a table with columns Subject ID and SSM File. The subject ID column will be automatically filled with all subject ID's detected in the loaded multi-subject SMP file (e.g. "AA", "MR"... in the used example).
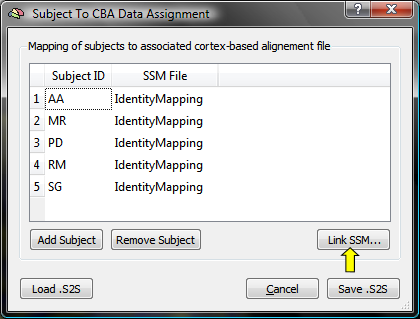
The SSM File column is filled at the beginning with entry "IdentityMapping" indicating that no CBA transformation file has been specified for the respective subject. To associate a subject ID with the subject's SSM file, highlight the row of a subject and then click the Link .SSM button (see snapshot above, yellow arrow); in the appearing Open File dialog, browse to the SSM file for this subject, which has been created earlier when running the CBA procedure. Note that the SSM file must correspond to the hemisphere currently used ("LH" or "RH"); the creation of the association table is typically performed separately for each cortical hemisphere (except in case that the subject's SPH meshes and SSM files are merged into one data structure). If the table had been previously already created and saved to disk, it may be reloaded using the Load .S2Sbutton. The dialog also allows to add additional subjects by using the Add Subject button or to remove a subject and the associated information by highlighting the subject's row and clicking the Remove Subject button; these options are generally not needed (and recommended), but may be useful if a table for a broader (or smaller) group of subjects will be created than the one specified in the multi-subject SMP file.
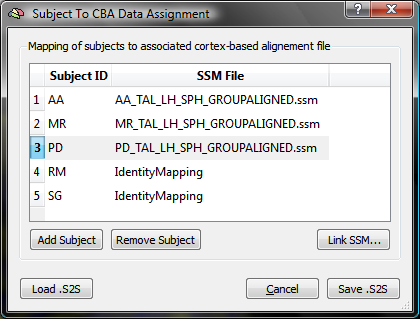
The snapshot above shows the state of the table after the SSM files for three (out of five) subjects have been assigned to the respective subject identifiers for the used example data set.
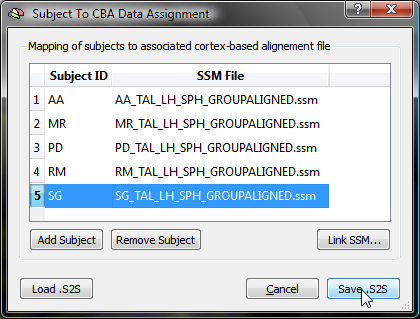
When all SSM files have been linked to the corresponding subject ID's, the table can be saved to disk by clicking the Save .S2S button. After having specified a name for the created table (e.g. "FiveSubjects_LH.s2s" in this example), the Subject To CBA Data Assignment dialog closes. The S2S File text field in the Apply cb-alignment to maps created in subject space field in the Surface Map Options dialog will automatically be filled with the saved .S2S file name (see snapshot below). Note that when other surface maps of the same group of subjects (and for the same hemisphere) must be cb-aligned, the saved .S2S file can be specified directly by using the Browse S2S File button, i.e. without using the Subject To CBA Data Assignment dialog.
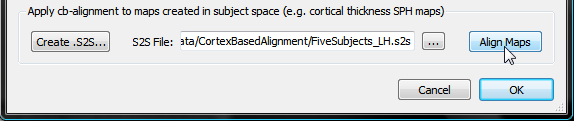
The surface maps of the subjects can now be transformed into group-aligned space by clicking the Align Maps button in the Apply cb-alignment to maps created in subject space field (see snapshot above). The resulting aligned surface maps are automatically saved to disk using the name of the original SMP with an added "_ALIGNED" string (see snapshot below, red arrow) allowing to easily distinguish the non-aligned from the cb-aligned surface maps.
To visualize how sulci and gyri overlap in group-aligned space for the used example data, the maps of all five subjects are again overlaid on the folded group brain (left hemisphere). The snapshot below shows that the cb-aligned curvature maps are now overlapping much better than the unaligned surface curvature maps.
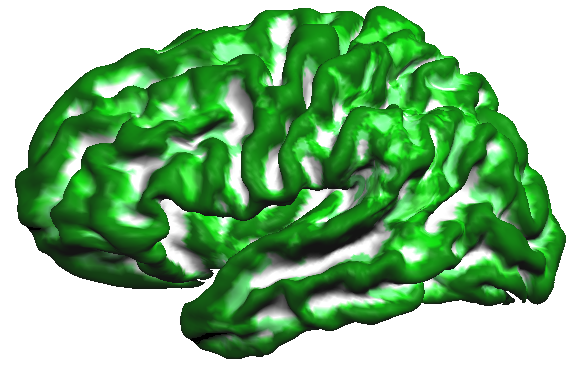
This observation is confirmed when creating a probabilistic map from the cb-aligned curvature maps. The probabilistic map uses binarized values, i.e. positive values indicating concave curvature/sulci are set to "1" while negative values indicating convex curvature/gyri are set to "0" in the surface map of a single subject. When sulci and gyri would be perfectly aligned across subjects, a probabilistic map should, thus, show the highest values (100% = 5 x "1") for sulci and the lowest values (0% = 5 x "0") for gyri. The calculated probabilistic curvature map comes quite close to this predicted ideal outcome as shown in the snapshot below (white = 100% overlap, dark green = 0% overlap).
Copyright © 2014 Rainer Goebel. All rights reserved.